Restriction enzymes cleave DNA at particular short sequences.
In addition to amplifying segments of DNA, researchers often also make use of techniques that cut DNA at specific sites. Cutting DNA molecules allows pieces from the same or different organisms to be brought together in recombinant DNA technology, which is discussed in the next section. It also is a way to determine whether or not specific sequences are present in a segment of DNA, as techniques for cutting DNA depend on specific DNA sequences. Finally, cutting DNA allows whole genomes to be broken up into smaller pieces for further analysis, such as DNA sequencing.
The method for cutting DNA makes use of a class of enzyme that recognizes specific, short nucleotide sequences in double-
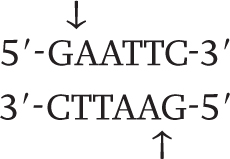
Wherever the enzyme finds this site in a DNA molecule, it cleaves each strand exactly at the position indicated by the vertical arrows. Note that the EcoRI restriction site is symmetrical: Reading from the 5′ end to the 3′ end, the sequence of the top strand is exactly the same as the sequence of the bottom strand. This kind of symmetry is palindromic (it reads the same in both directions) and is typical of restriction sites. Note also that the site of cleavage is not in the center of the recognition sequence. The cleaved double-
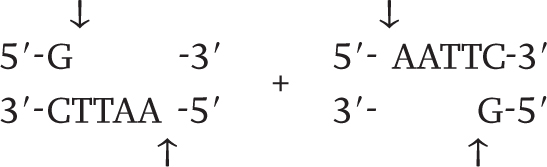
261
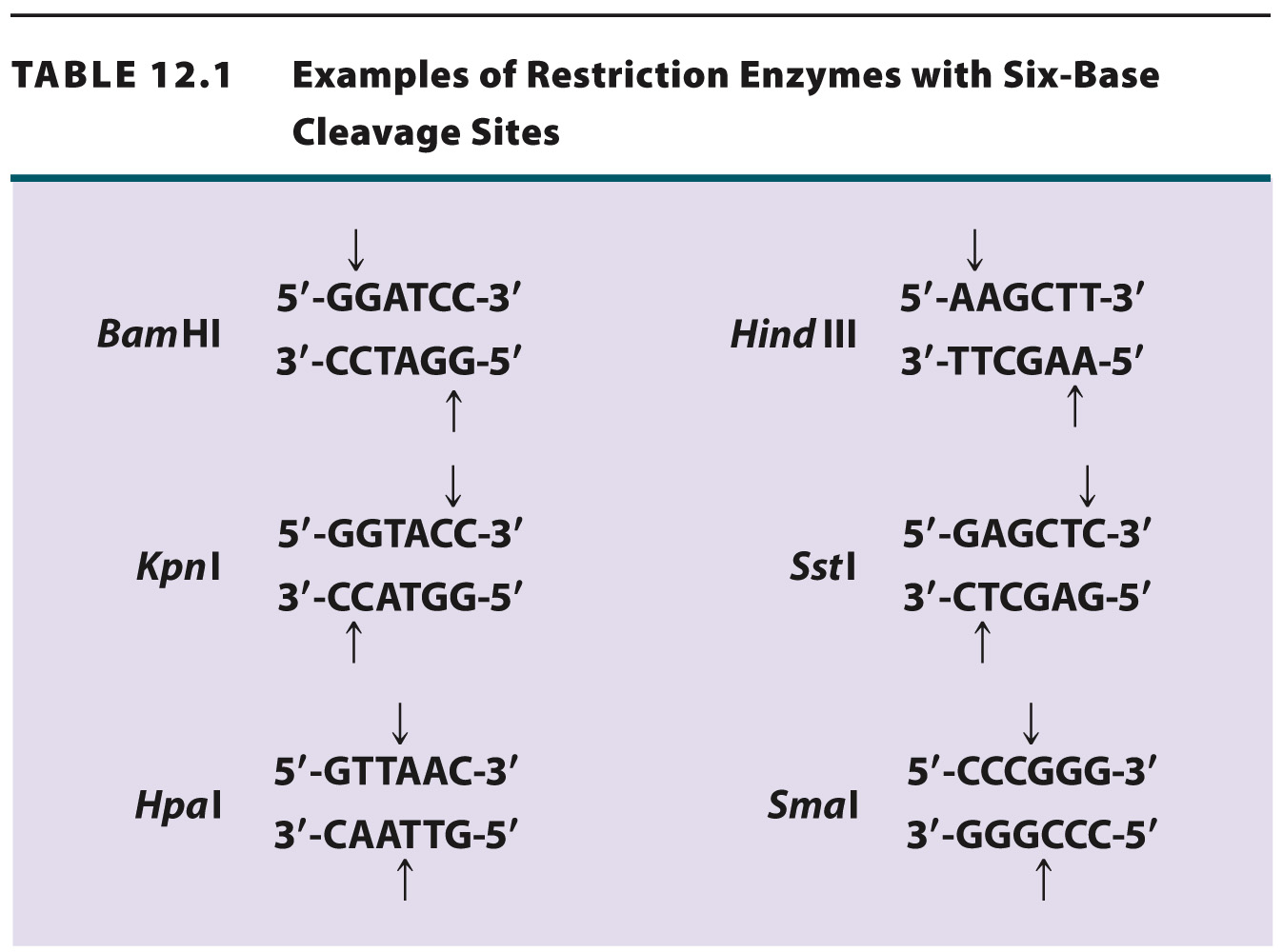
Table 12.1 shows more examples of restriction enzymes. Some cleave their restriction site to produce a 5′ overhang, others produce a 3′ overhang, and still others cleave in the middle of their restriction site and leave blunt ends with no overhang. The standard symbols for restriction enzymes include both italic and roman letters. The italic letters stand for the species from which the enzyme is derived (E. coli in the case of EcoRI, Bacillus amyloliquefaciens in the case of BamHI), and the Roman letters designate the particular restriction enzyme isolated from that species.
When a particular restriction enzyme is used to break up a whole genome into smaller fragments, the specificity of restriction enzymes ensures that the DNA from each cell in an individual organism yields the same set of fragments and any particular DNA sequence present in the cells is contained in a fragment of the same size. If the genome is small enough that the number and sizes of the fragments are limited, the individual fragments can be visualized directly in a gel (Fig. 12.15). These fragments can then be extracted from the gel for further analysis or manipulation. For instance, the extracted fragments can be sequenced or ligated to other fragments.
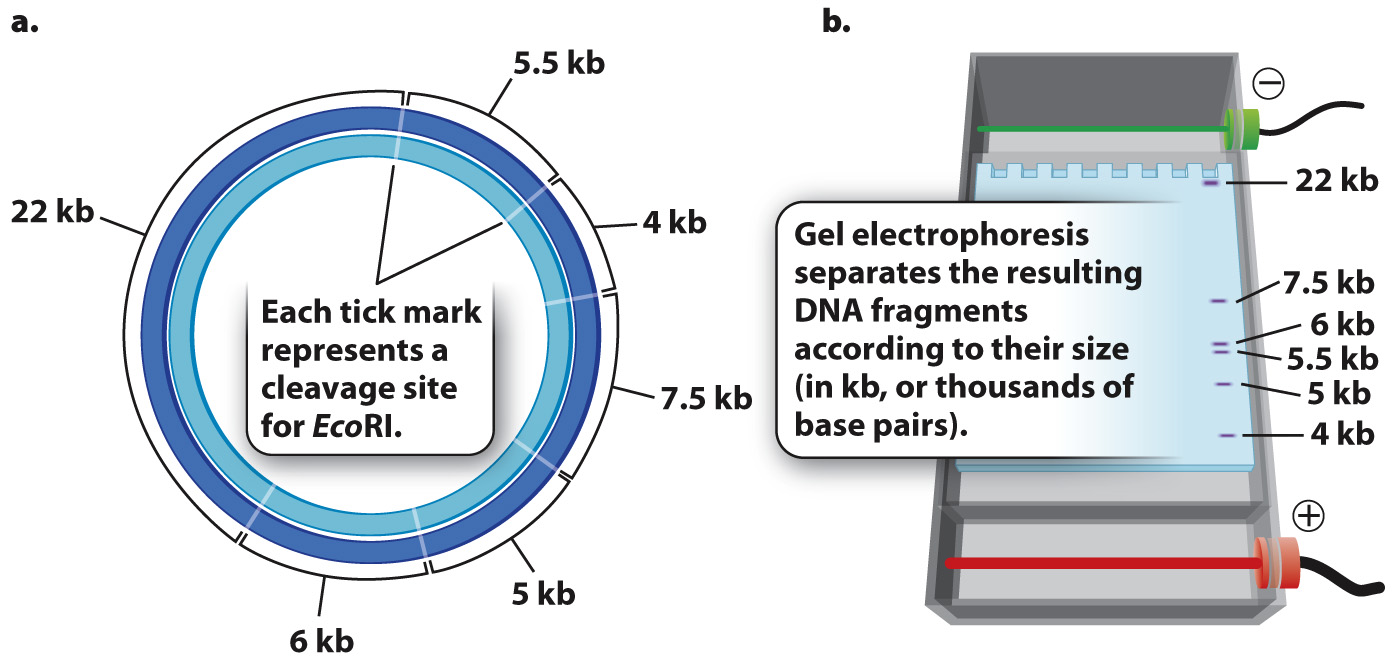
But most genomes are too large to give such a simple picture. For example, the human genome is cleaved into more than a million different fragments by EcoRI, and these fragments appear as one big smear in a gel rather than as a series of discrete bands. In the next section, we discuss how individual bands containing a sequence of interest can be detected in such a gel.