DNA strands can be separated and brought back together again.
A researcher may wish to know whether a gene in one species is present in the DNA of a related species, but the nucleotide sequence of the gene is unknown. In such a case, techniques like PCR that require knowledge of the target DNA sequence cannot be used. Instead, the researcher can determine whether a DNA strand containing the gene in one species can base pair with the complement of a similar gene in a DNA strand from the other species. The base pairing of complementary single-
Denatured DNA strands from one source can renature with DNA strands from a different source if their sequences are precisely or mostly complementary. Two very closely related sequences will have more perfectly matched bases and thus more hydrogen bonds holding them together than two sequences that are less closely related. The degree of base pairing between two sequences affects the temperature at which they renature: Very closely related sequences renature at a higher temperature than less closely related sequences. Evolutionary biologists have used this principle to estimate the proportion of perfectly matched bases present in the DNA of different species, which is used as a measure of how closely those species are related. In general, the more closely two species are related, the more similar their DNA sequences. Knowledge of species relatedness is important in many applications, including conservation, identifying endangered species, and tracing the evolutionary history of organisms.
Renaturation makes it possible to use a small DNA fragment as a probe. This fragment is usually attached to a light-
Let’s say you are interested in determining the number of copies of a particular DNA sequence in a genome or determining whether a given gene in human genomic DNA is intact. Recall that human and other genomic DNA cut with a restriction enzyme yield a large number of DNA fragments that look like a smear following gel electrophoresis. A labeled probe can be used to determine the size and number of a DNA sequence of interest in a gel. The method is known as a Southern blot (Fig. 12.16) after its inventor, the British molecular biologist Edwin M. Southern.
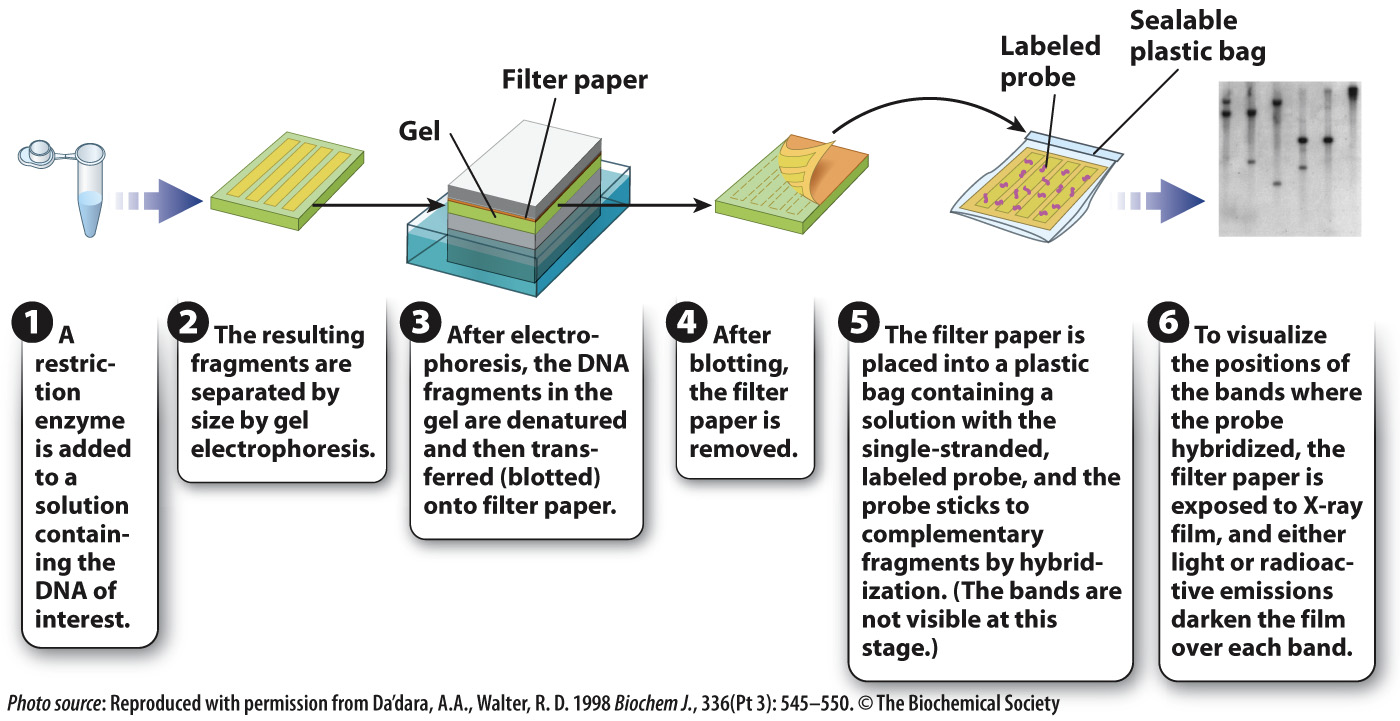
In a Southern blot, single-
The presence of a band or bands on the film therefore indicates the number and size of DNA fragments that are complementary to the probe. This information in turn tells you the sizes and the number of copies of a particular DNA sequence present in the starting sample.