Synthesis of the leading and lagging strands is coordinated.
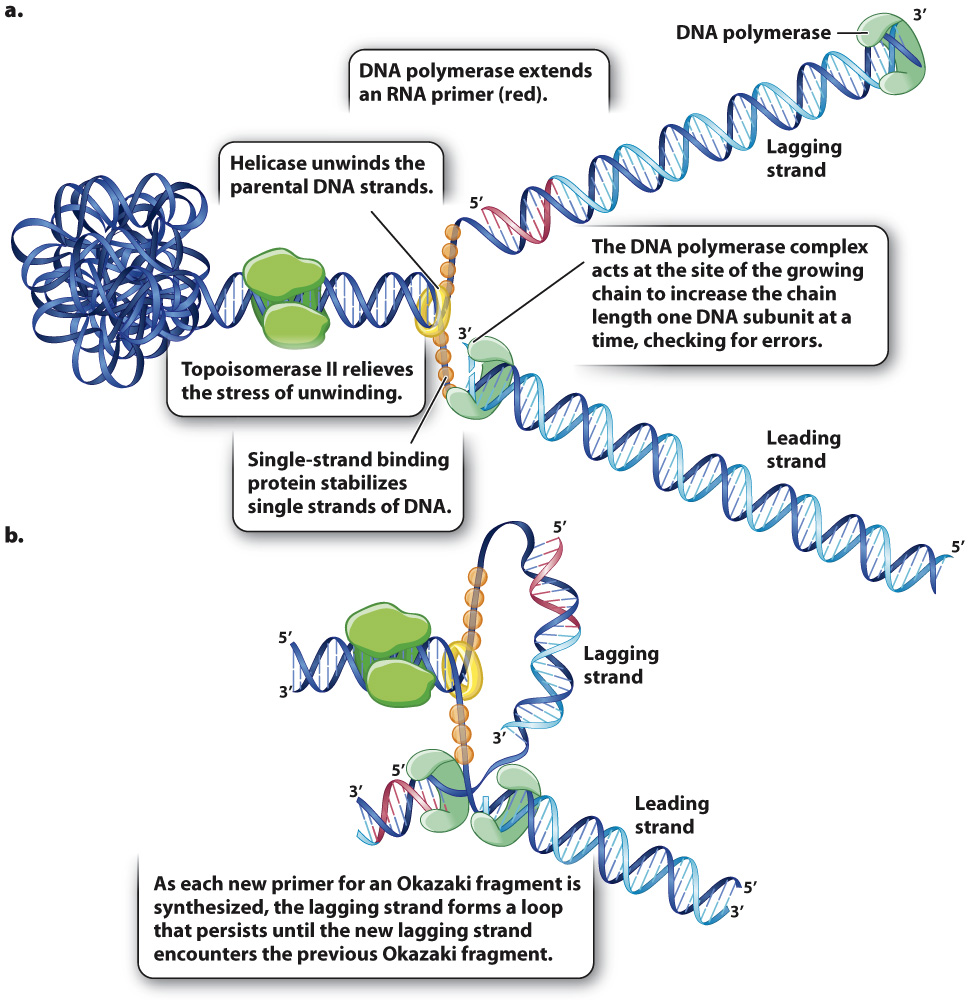
Fig. 12.7a shows an overview of DNA replication including the DNA polymerase complex that elongates each strand at the 3′ end, the RNA primase that makes the primer, the DNA polymerase complex that replaces the RNA primer with DNA, and the DNA ligase that joins the DNA fragments in the lagging strand. Many other proteins and enzymes work at the same time to ensure accurate and efficient synthesis of the daughter strands. One of these, topoisomerase II, works upstream from the replication fork to relieve the stress on the double helix that results from its unwinding at the replication fork. In the meantime, helicase separates the strands of the parental double helix at the replication fork. Then, single-
253
While Fig. 12.7a makes it clear how synthesis of the leading strand and the lagging strand take place, it does not show how synthesis of these strands occurs at the same time and rate. To help ensure that both strands of the double helix are replicated at nearly the same rate, synthesis of the leading and lagging strands is coordinated as shown in Fig. 12.7b. The DNA polymerase complexes for each strand stay in contact with each other, with the lagging strand’s polymerase releasing and retrieving the lagging strand for the synthesis of each new RNA primer. The positioning of the polymerases is such that both the leading strand and the lagging strand pass through in the same direction, which requires that the lagging strand be looped around as shown in Fig. 12.7b. In this configuration, the 3′ end of the lagging strand and the 3′ end of the leading strand are elongated together, which ensures that neither strand outpaces the other in its rate of synthesis.
254
As we will see in Chapter 14, when DNA damage occurs during replication, the rate of synthesis slows down so that the DNA can be repaired. If synthesis of one strand slows down to repair DNA damage, synthesis of the other strand slows down, too. The pairing of the replication complexes is disrupted when the RNA primer of the previous Okazaki fragment is encountered, and then takes place again when a new lagging-