The HIV genome illustrates the utility of genome annotation and comparison.
HIV and related viruses provide an example of how genomes are annotated and how comparisons among genome sequences can reveal evolutionary relationships. A virus is a small infectious agent that contains a nucleic acid genome packaged inside a protein coat called a capsid. Viruses can bind to receptor proteins on cells of the host organism, enabling the viral genome to enter the cell. Infection of a host cell is essential to viral reproduction because viruses use cellular ATP and hijack cellular machinery to replicate, transcribe, and translate their genome in order to make more viruses. Later in this chapter we will see that viruses can differ in the types of nucleic acid that make up their genome and how their messenger RNA is produced, but here we consider only the genome of HIV.
Whereas the genome of all cells consists of double-
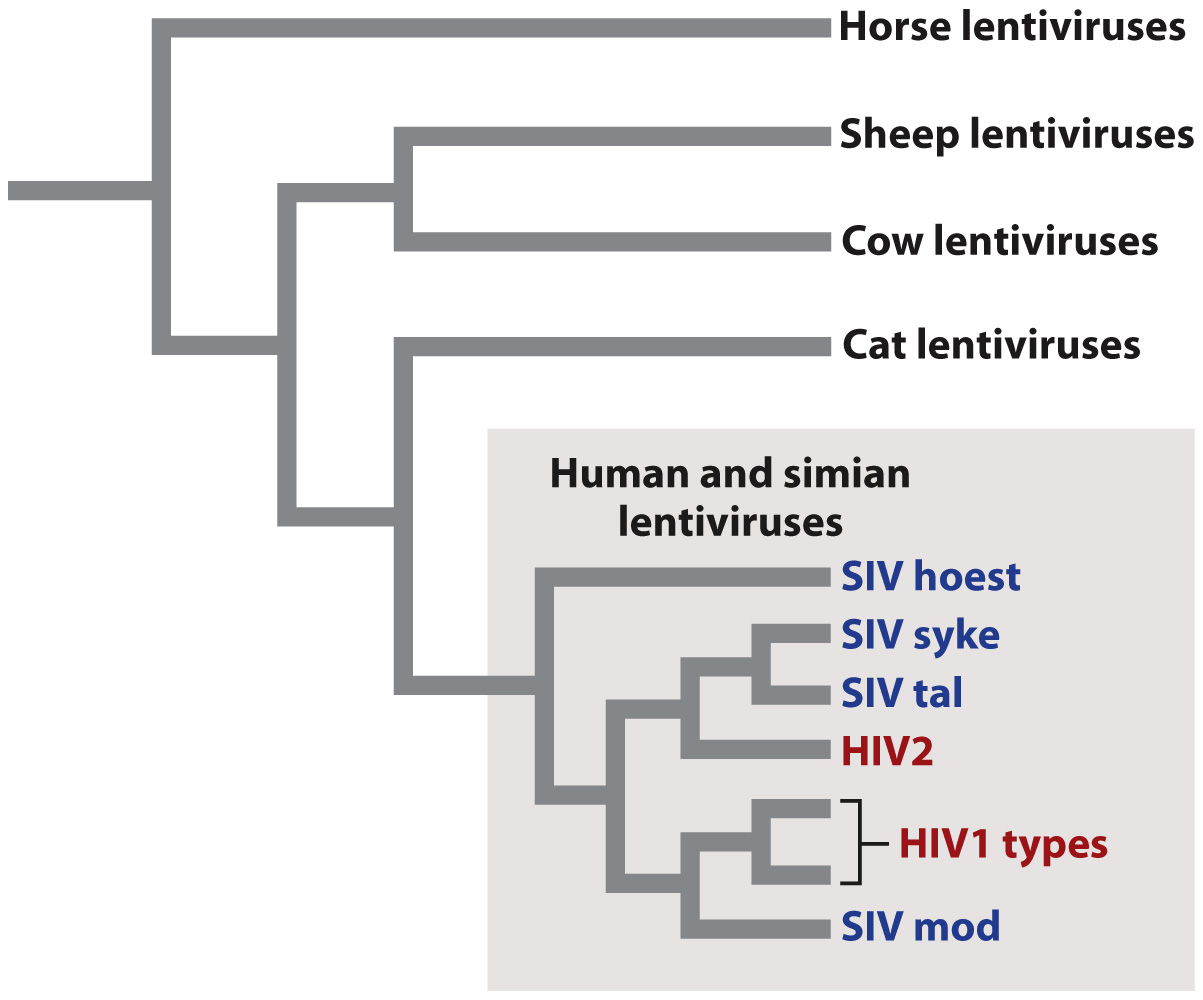
Fig. 13.6 shows the evolutionary relationships among a sample of lentiviruses, grouped according to the similarity of their genome sequences. The evolutionary tree shows that closely related viruses have closely related hosts. For example, simian lentiviruses are more closely related to one another than they are to cat lentiviruses. This observation implies that the genomes of the viruses evolve along with the genomes of their hosts. A second feature shown by the evolutionary tree is that human HIV originated from at least two separate simian viruses that switched hosts from simians (most likely chimpanzees) to humans.
278
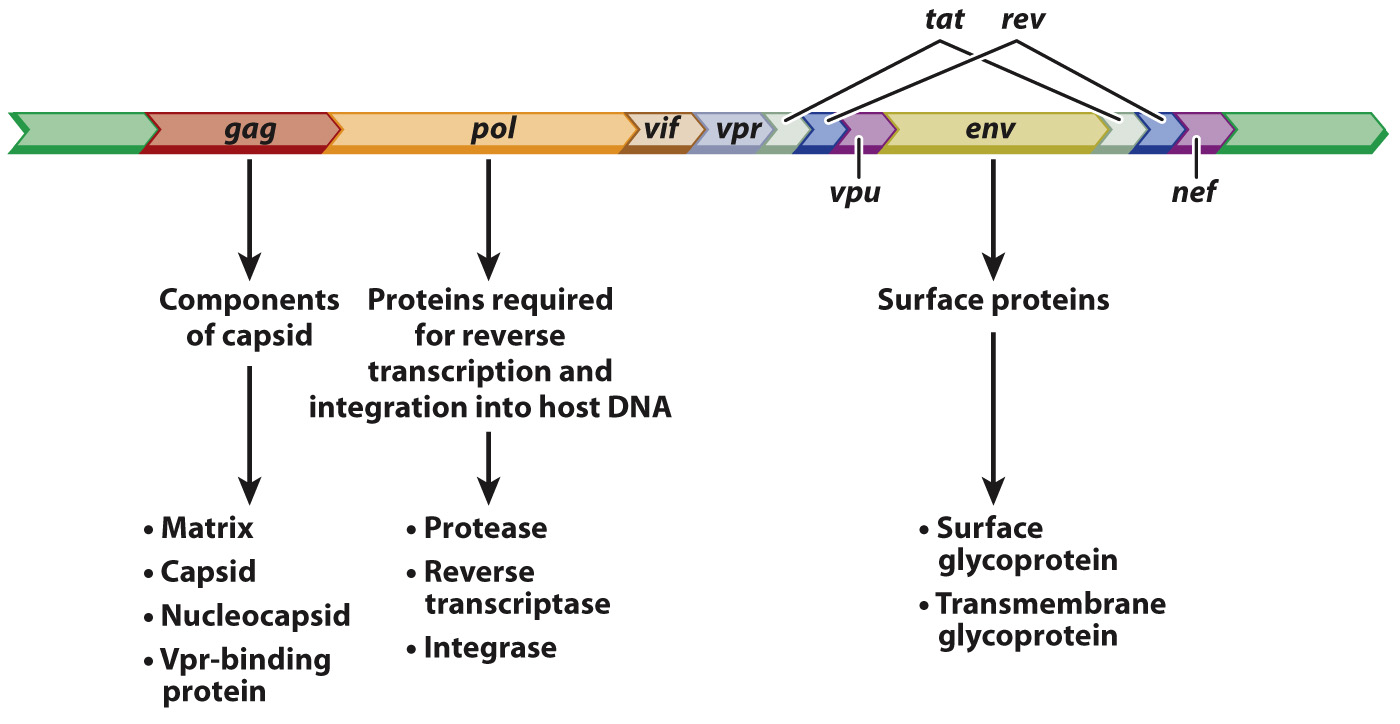
The annotated sequence of the HIV genome tells us a lot about the biology of this virus (Fig. 13.7). Many details are left out here, but the main point is to show the functional elements of HIV in the form of an annotated genome. The open reading frame denoted gag encodes protein components of the capsid, pol encodes proteins needed for reverse transcription of the viral RNA into DNA and incorporation into the host genome, and env encodes proteins that are embedded in the lipid envelope. The annotation in Fig. 13.7 also includes the genes tat and rev, encoding proteins essential for the HIV life cycle, as well as the genes vif, vpr, vpu, and nef, which encode proteins that enhance virulence in organisms. Identification of the genes necessary to complete the HIV cycle is the first step to finding drugs that can interfere with the cycle and prevent infection.
Quick Check 3 Fig. 13.6 shows that closely related lentiviruses have closely related host organisms. What does this observation suggest about the ability of lentiviruses to infect a wide variety of hosts?
Quick Check 3 Answer
The observation suggests that lentiviruses and their hosts evolve together and are closely matched. This in turn suggests that lentiviruses are usually unable to infect hosts that are not evolutionarily very closely related to their natural host.