Eukaryotic cells package their DNA as one molecule per chromosome.
In eukaryotic cells, DNA in the nucleus is packaged differently from DNA in bacteria. As discussed in Chapter 3, eukaryotic DNA is linear and each DNA molecule forms a single chromosome. In a chromosome, DNA is packaged with proteins to form a DNA-
This first level of packaging of the DNA is sometimes referred to as “beads on a string,” with the nucleosomes the beads and the DNA the string. It is also called a 10-
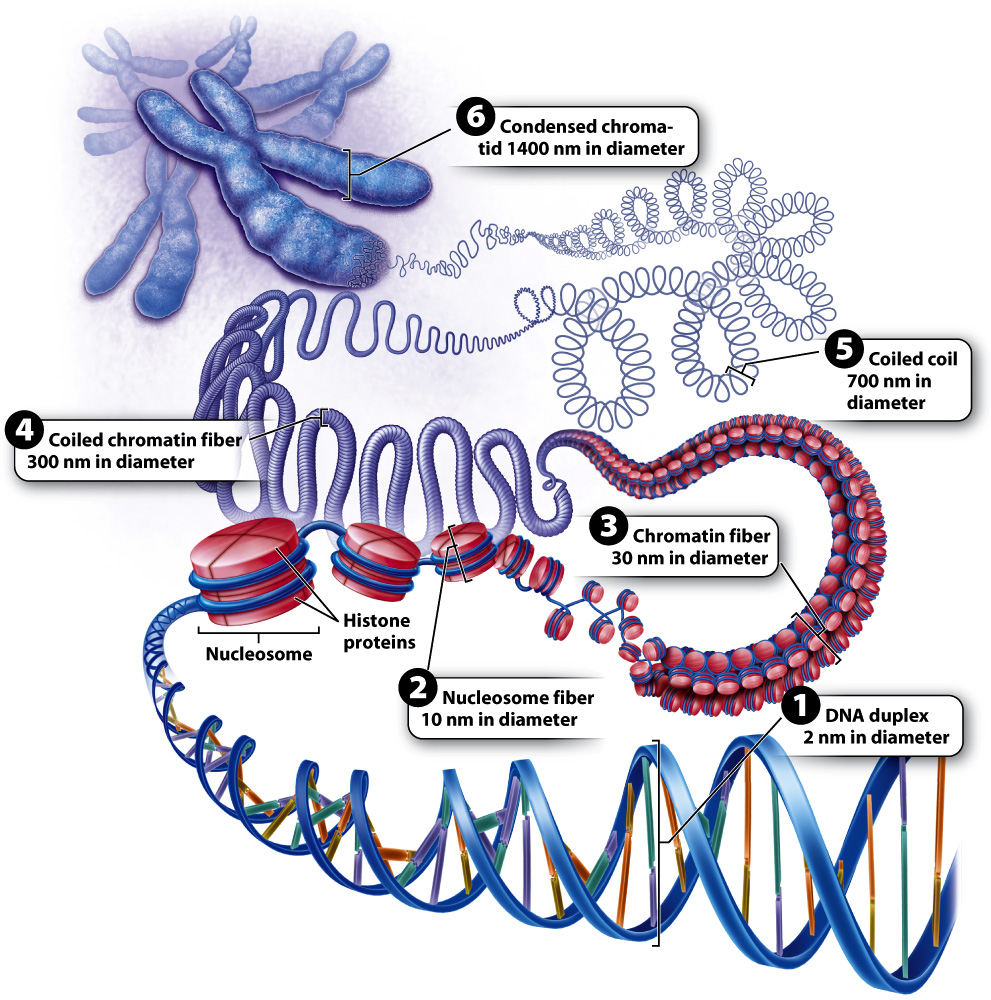
The next level of packaging occurs when the chromatin is more tightly coiled, forming a 30-
Greater detail of the structure of a fully condensed chromosome is revealed when the histones are chemically removed (Fig. 13.13). Without histones, the DNA spreads out in loops around a supporting protein structure called the chromosome scaffold. Each loop of relaxed DNA is 30 to 90 kb long and anchored to the scaffold at its base. Before removal of the histones, the loops are compact and supercoiled. Each human chromosome contains 2000–
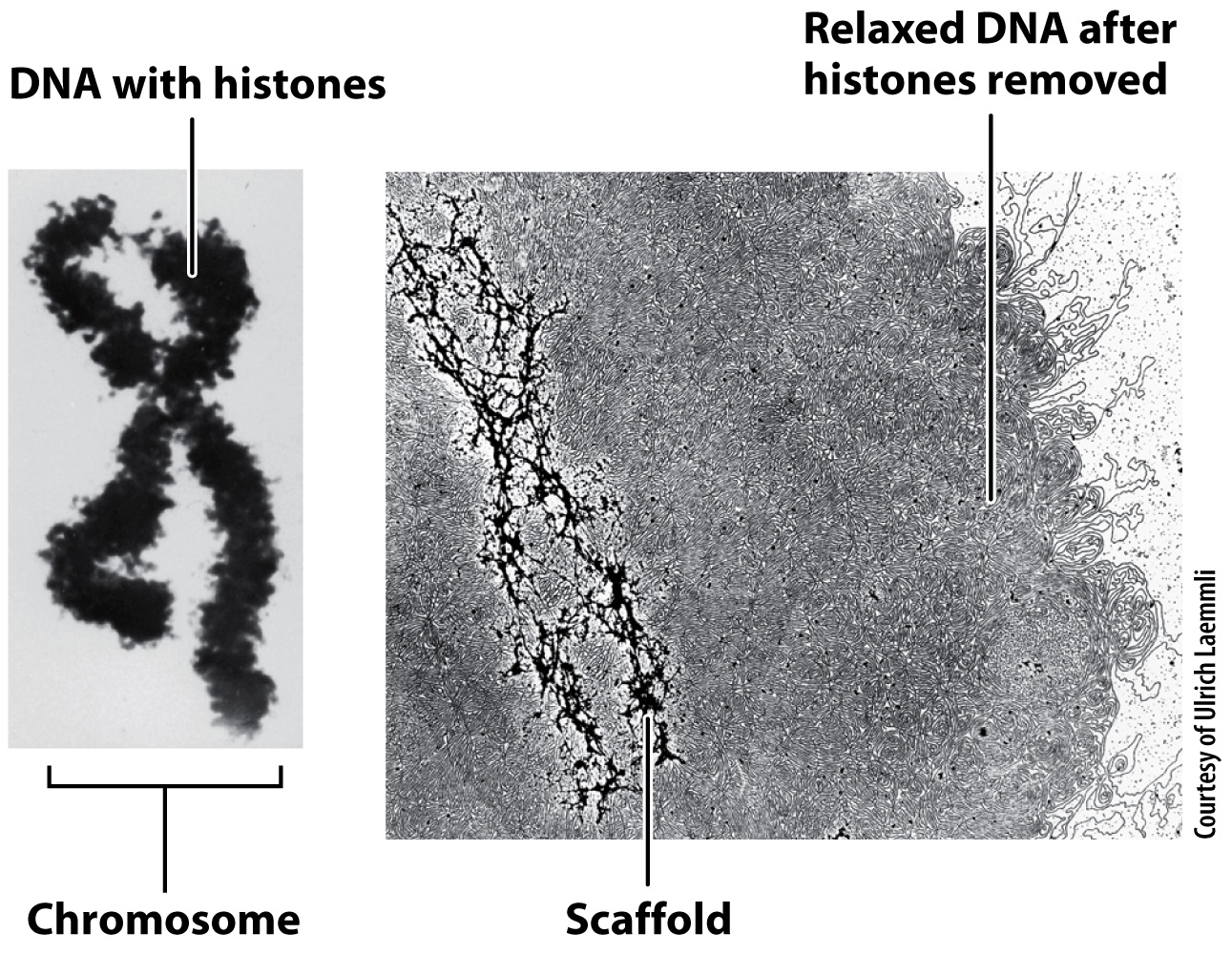
Despite intriguing similarities between the nucleoid model in Fig. 13.11 and the chromosome scaffold model in Fig. 13.13, the structures evolved independently and make use of different types of protein to bind the DNA and form the folded structure of DNA and protein. Furthermore, the size of the eukaryotic chromosome is vastly greater than the size of the bacterial nucleoid. To appreciate the difference in scale, keep in mind that the volume of a fully condensed human chromosome is five times larger than the volume of a bacterial cell.