Most DNA damage is corrected by specialized repair enzymes.
Cells contain many specialized DNA-
As we saw earlier in this chapter, mispairing of bases during DNA replication leads to the incorporation of incorrect nucleotides and potentially to nucleotide substitutions. About 99% of the mispaired bases are corrected immediately by the proofreading function of DNA polymerase, in which the mispaired nucleotide is removed immediately after incorporation and replaced by the correct nucleotide (Chapter 12).
Even after proofreading by DNA polymerase, the proportion of mismatched nucleotides in replicated DNA is about 10–6, which is at least 1000 times greater than the actual frequency of errors in cellular organisms (see Fig. 14.1). What accounts for the difference is a second-
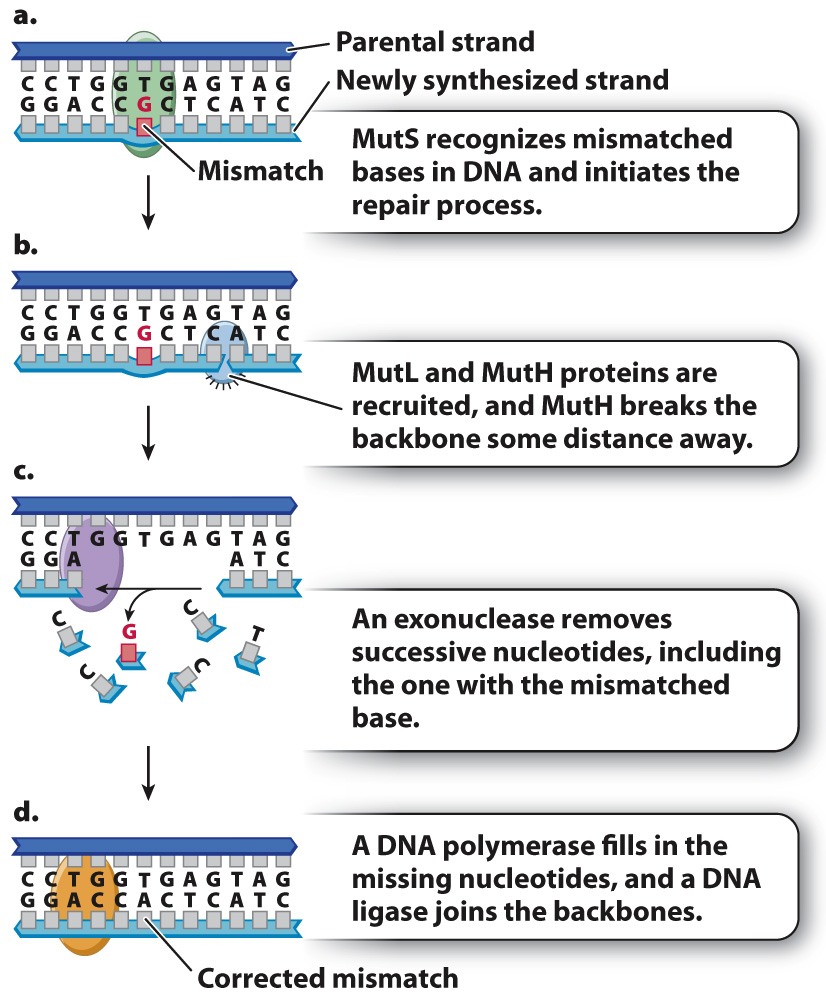
In the bacterium E. coli, postreplication mismatch repair begins when a protein known as MutS binds to the site of a mismatch (Fig. 14.17a) and brings two other proteins (MutL and MutH) to the site. MutL determines which DNA backbone will be cleaved, and MutH cleaves the backbone in the vicinity of the mismatch (Fig. 14.17b). An exonuclease degrades the cleaved strand to a distance beyond the site of the mismatch (Fig. 14.17c). A DNA polymerase then synthesizes new DNA to fill the gap, correcting the mismatch, and a DNA ligase seals the remaining nick in the DNA backbone. Postreplication mismatch repair usually, but not always, cleaves the daughter strand, so the corrected strand matches the parental template strand.
305
Eukaryotes have proteins corresponding to those in Fig. 14.17, and the importance of postreplication mismatch repair is underlined by the finding that a genetic predisposition to certain types of colon cancer results from mutations in the human counterparts of MutS and MutL.
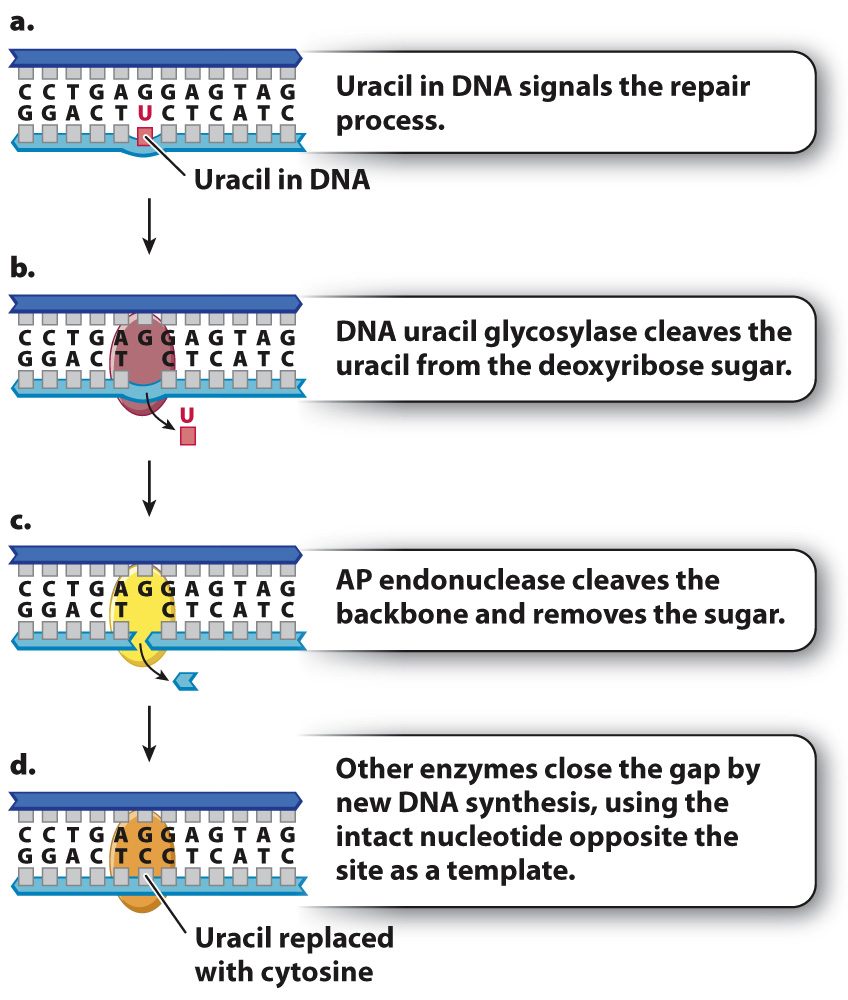
The result of proofreading by DNA polymerase and postreplication mismatch repair is an increase in the fidelity of DNA replication. Cells have evolved many other systems that repair different types of DNA damage that can occur even in nondividing cells. One example is base excision repair, which corrects abnormal or damaged bases. In the first step of base excision repair, an abnormal or damaged base is cleaved from the sugar in the DNA backbone. Then, the baseless sugar is removed from the backbone, leaving a gap of one nucleotide. Finally, a repair polymerase inserts the correct nucleotide into the gap.
An example of base excision repair is shown in Fig. 14.18, which depicts a molecule in which uracil instead of cytosine is present in a DNA strand (Fig. 14.18a). Recall that uracil is normally found in mRNA, but not in DNA. Most organisms have an enzyme called DNA uracil glycosylase that cleaves the uracil from its deoxyribose sugar, resulting in a sugar in the DNA backbone lacking a base (Fig. 14.18b). Such baseless sugars are recognized by an enzyme called AP endonuclease (Fig. 14.18c), which cleaves the backbone on both sides of the sugar that lacks a base. This cleavage results in a one-
306
Cells have also evolved mechanisms to repair short stretches of DNA containing mismatched or damaged bases. One of these is known as nucleotide excision repair, which has a similar mechanism of action to mismatch repair but uses different enzymes (Fig. 14.19). Instead of degrading a DNA strand nucleotide by nucleotide until the mismatch is removed, nucleotide excision repair removes an entire damaged section of a strand at once. Nucleotide excision repair is also used to remove nucleotides with bulky side groups, as well as thymine dimers resulting from ultraviolet light.
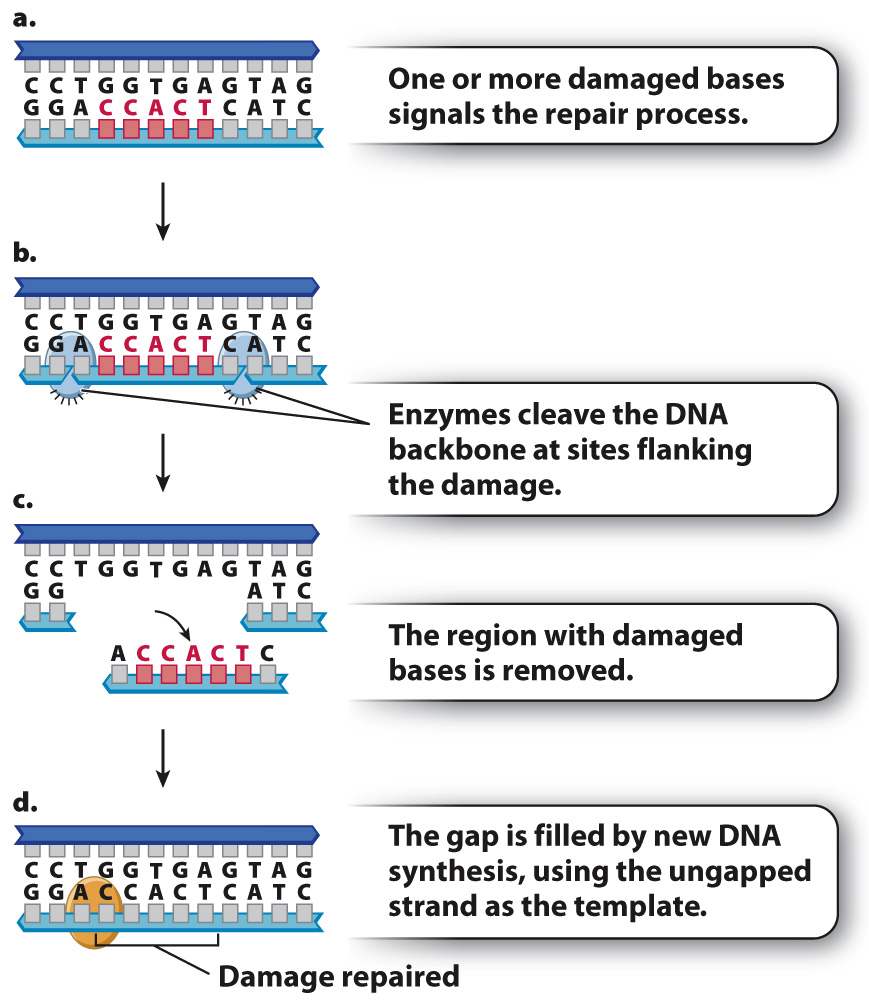
The importance of excision repair is illustrated by the disease xeroderma pigmentosum (XP), in which nucleotide excision repair is defective. People with XP are exquisitely sensitive to the UV radiation in sunlight. Because of the defect in nucleotide excision repair, damage to DNA resulting from UV light is not corrected, leading to the accumulation of mutations in skin cells. The result is high rates of skin cancer. People with XP must minimize their exposure to sunlight and in extreme cases stay out of the sun altogether.
When DNA repair mechanisms function properly, they reduce the rate of mutation to a level that is compatible with life. But they are not perfect, and they do not catch all the mistakes. The result is genetic variation, the raw material of evolution.