Single-nucleotide polymorphisms (SNPs) are single-base changes in the genome.
One of the most common types of genetic variation is a difference in a nucleotide at a specific site. A single-nucleotide polymorphism (SNP) is a site in the genome where either of two different nucleotide pairs can occur and where each nucleotide pair is common enough in the population to be present in a random sample of 50 diploid individuals. The differences among the hemoglobin alleles are SNPs. The A, S, and C alleles differ from one another at just one nucleotide site (see Fig. 15.1).
Eye color is also associated with a SNP. The blue eye phenotype results from reduced expression of a gene called OCA2, which encodes a membrane protein involved in the transport of small molecules, including the amino acid tyrosine, which is a precursor of the melanin pigment associated with the brown eye phenotype. Although more than a dozen alleles of OCA2 are known that have an amino acid replacement in the protein, none of these results in blue eyes.
The SNP implicated in blue eyes is a site that is a C–G base pair in one allele. The other common allele has a T–A at this position, which is not associated with blue eyes (Fig. 15.7). In one study, the homozygous C–G genotype was found in 94% of 183 individuals with blue eyes and in only 2% of 176 individuals with brown eyes. This strong association is quite unexpected because the SNP associated with blue eyes is in an intron of a neighboring gene, and is therefore noncoding. The proposed mechanism for the association is that the C–G allele makes the adjacent OCA2 gene less accessible to transcription factors, thereby reducing the amount of the transporter protein and consequently the production of melanin in the iris. The proposed mechanism may be right or wrong; this example emphasizes that scientists have much to learn about the possible phenotypic effects of noncoding DNA.
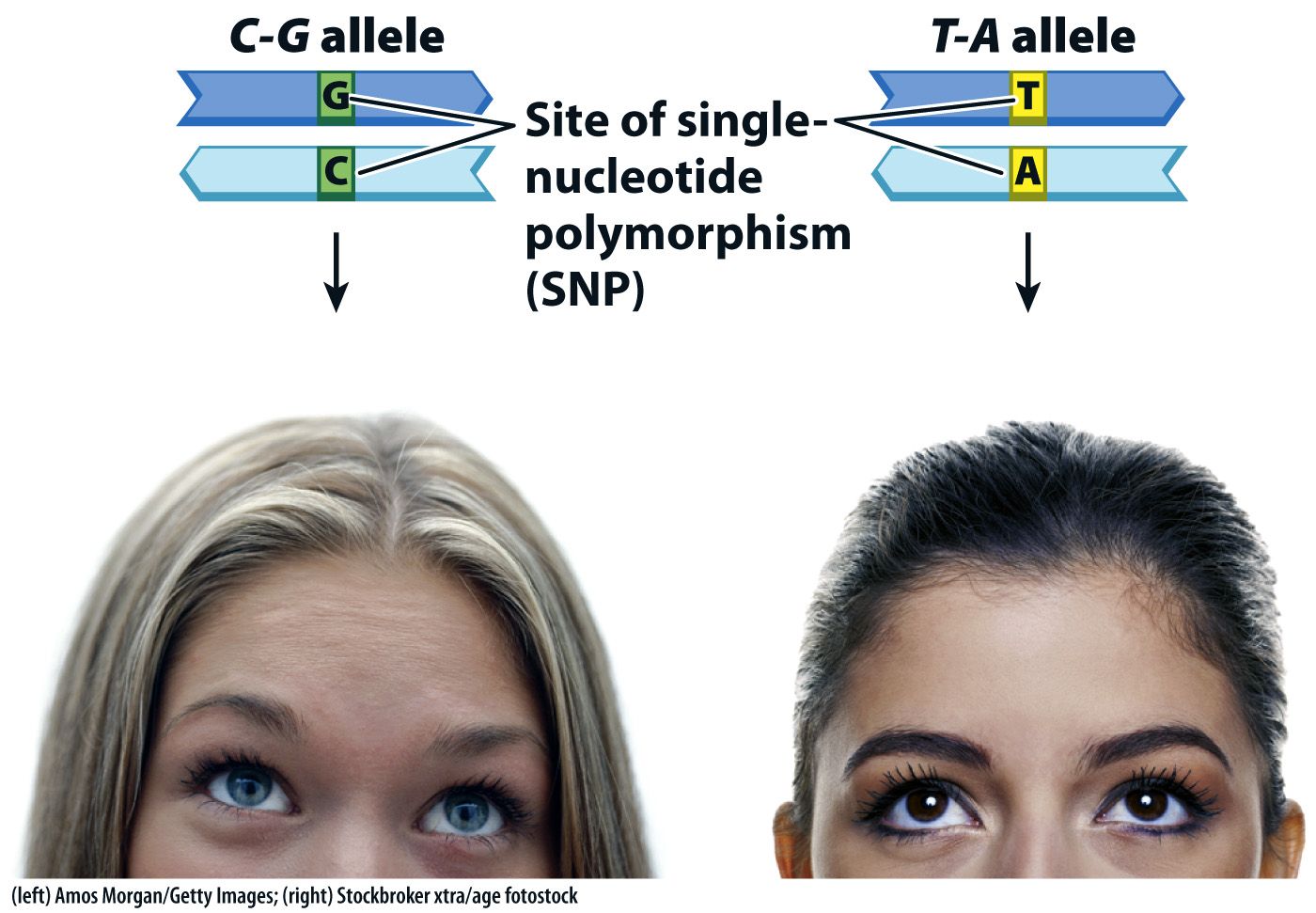
FIG. 15.7 Single-nucleotide polymorphisms (SNPs). A SNP in the region neighboring OCA2 is strongly associated with blue eyes.
The C–G versus T–A SNP is typical of most SNPs in that only two of the four possible base pairs (G–C and A–T in addition to C–G and T–A) are present in the population at any appreciable frequency.
Quick Check 5 What’s the difference between a point mutation and a SNP?
Quick Check 5 Answer
A point mutation is a change in a single nucleotide in an individual cell, such as C to G. An SNP results from a point mutation that occurred at some time in the past so that now, in the population, there are two or more different single nucleotides at a given position. For example, some people might have C at a certain position and others might have G at that position.
