Nearby genes in the same chromosome show linkage.
Genes that are sufficiently close together in the same chromosome are said to be linked. That is, they tend to be transmitted together in inheritance and do not assort independently of each other as Mendel observed. Note that linked genes refer to two genes that are close together in the same chromosome, which may be an autosome or sex chromosome. This is not to be confused with an X-
Linkage was discovered in Drosophila by Alfred H. Sturtevant, another of Morgan’s students. Once again, genes in the X chromosome played a key role in the discovery because the phenotype of the males reveals their genotype as in a testcross with an autosomal recessive. An example is shown in Fig. 17.9. Sturtevant worked with male fruit flies that have an X chromosome carrying two recessive mutations. One is in the white gene (w) discussed earlier, which when nonmutant results in fruit flies with red eyes and when mutant results in fruit flies with white eyes. The other recessive mutation is in a gene called crossveinless (cv), which when nonmutant results in fruit flies with tiny crossveins in the wings and when mutant results in the absence of these crossveins.
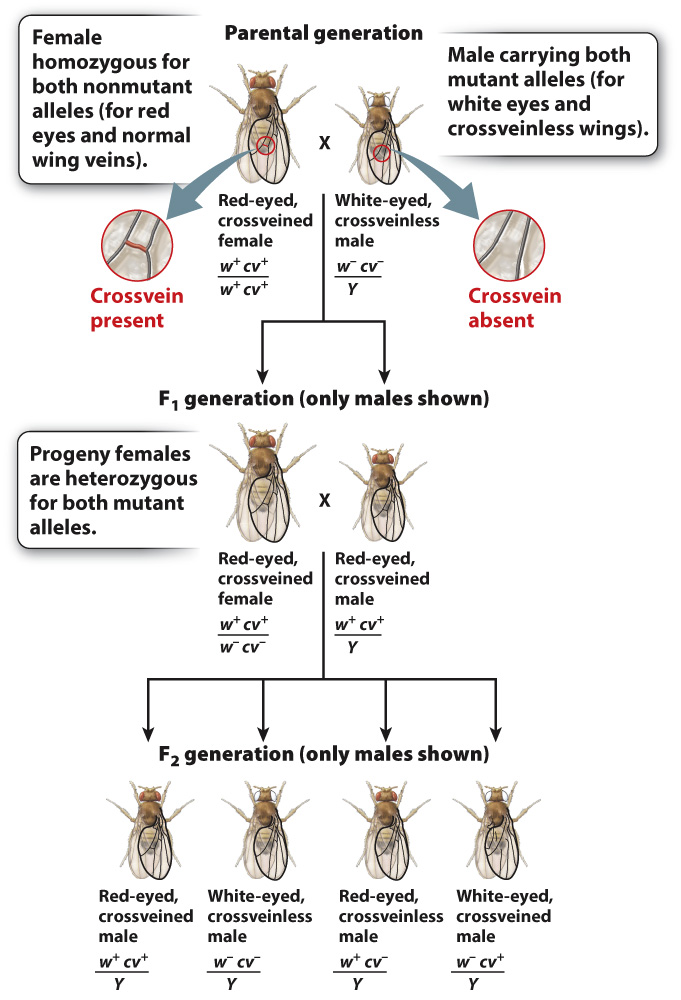
Sturtevant crossed this doubly mutant male with a female carrying the nonmutant forms of the genes (w+ and cv+) in both X chromosomes. He saw that the offspring consist of phenotypically wild-
354
As shown in Fig. 17.9, the male F2 progeny consist of four types:
| Genotype of F2 Progeny | Number of Fruit Flies |
| w+cv+/Y (red eyes, normal crossveins) | 357 |
| w–cv–/Y (white eyes, missing crossveins) | 341 |
| w+cv–/Y (red eyes, missing crossveins) | 52 |
| w–cv+/Y (white eyes, normal crossveins) | 45 |
Although all four possible classes of maternal gametes are observed in the male progeny, they do not appear in the ratio 1:1:1:1 expected when gametes contain two independently assorting genes (Chapter 16). The lack of independent assortment means that the genes show linkage.
The male progeny fall into two groups. One group, represented by larger numbers of progeny, derives from maternal gametes containing either w+cv+ or w–cv–. These are called nonrecombinants because the alleles are present in the same combination as that in the parent. The other group of male progeny consists of w+cv– and w–cv+ combinations of alleles. These are called recombinants, and they result from a crossover, the physical exchange of parts of homologous chromosomes, which takes place in prophase I of meiosis (Chapter 11).
Crossing over is a key process in meiosis (Chapter 11). Most chromosomes have one or more crossovers that form between the homologous chromosomes as they pair and undergo meiosis. Human females average about 2.75 crossovers per chromosome pair, and human males average about 2.50 crossovers per chromosome pair. As noted earlier for the X and Y chromosomes, crossovers between homologous chromosomes are important mechanically because they help hold the homologs together so they can align properly at metaphase I and segregate to opposite poles at anaphase I.
Fig. 17.10 shows how recombinant chromosomes arise from crossing over between genes, using the hypothetical genes A and B. In a cell undergoing meiosis in which a crossover takes place between the genes, the allele combinations are broken up in the chromatids involved in the exchange, and the resulting gametes are AB, Ab, aB, and ab (Fig. 17.10a).
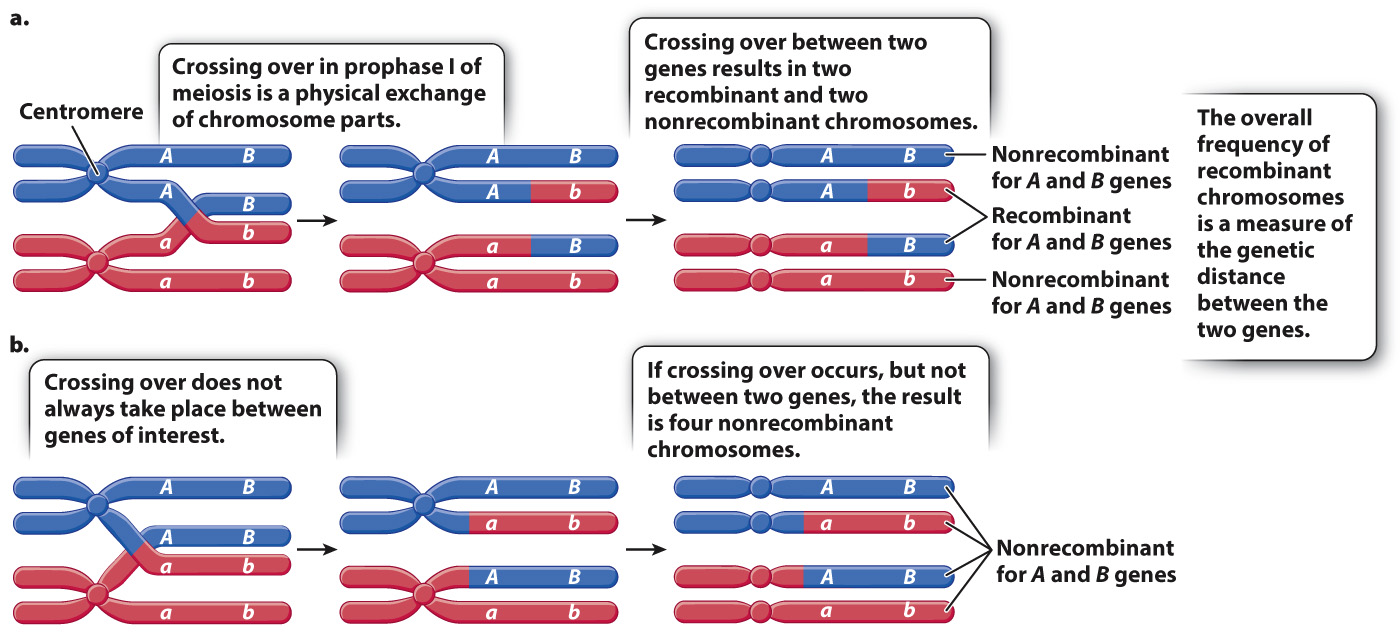
Note that crossing over does not result only in recombinant chromosomes. Fig. 17.10a shows that, even when a crossover occurs in the interval between the genes, two of the resulting chromosomes contain the nonrecombinant configuration of alleles. These nonrecombinant configurations occur because crossing over occurs at the four-
355
Fig. 17.10b shows a second way in which nonrecombinant chromosomes originate. When two genes are close together in a chromosome, a crossover may not occur in the interval between the genes but at some other location along the chromosome. In this example, the crossover occurs in the region between gene A and the centromere, and therefore the combination of alleles A and B remains intact in one pair of chromatids and the combination of alleles a and b remains intact in the homologous chromatids. The resulting gametes are equally likely to have either AB or ab, both of which are nonrecombinant.
Nonrecombinant chromosomes can also be the result of two crossover events occurring between two genes, in which the effects of the first crossover (creating recombinants) is reversed by a second crossover (re-