Gene expression can be influenced by chemical modification of DNA or histones.
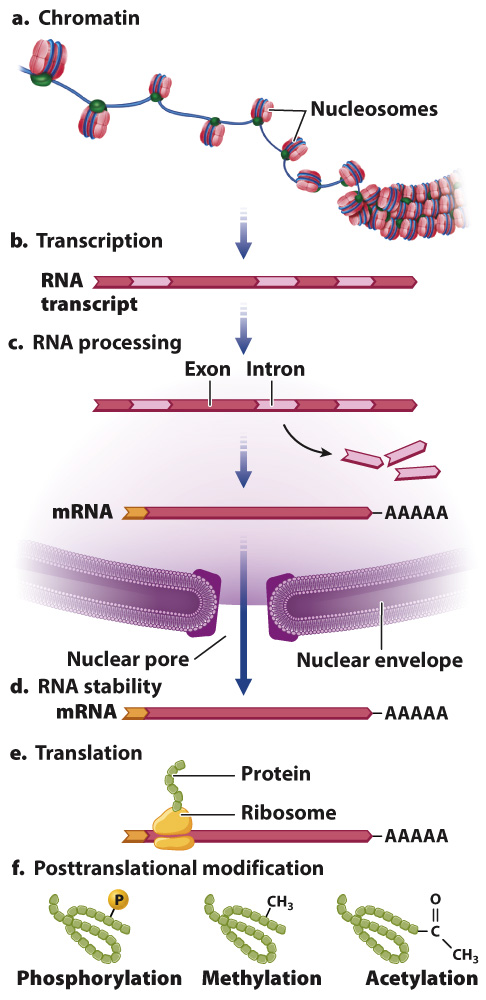
Fig. 19.1 shows the major places where gene regulation in eukaryotes can take place. The first level of control is at the chromosome, even before transcription takes place. The manner in which DNA is packaged in the nucleus in eukaryotes provides an important opportunity for regulating gene expression. Regulation at this level of gene expression is determined in part by whether the proteins necessary for transcription can gain physical access to the genes they transcribe.
DNA in eukaryotes is packaged as chromatin, a complex of DNA, RNA, and proteins that gives chromosomes their structure (Chapters 3 and 13). When chromatin is in its coiled state, the DNA is not accessible to the proteins that carry out transcription. The chromatin must loosen to allow space for transcriptional enzymes and proteins to work. This is accomplished through chromatin remodeling, in which the nucleosomes are repositioned to expose different stretches of DNA to the nuclear environment.
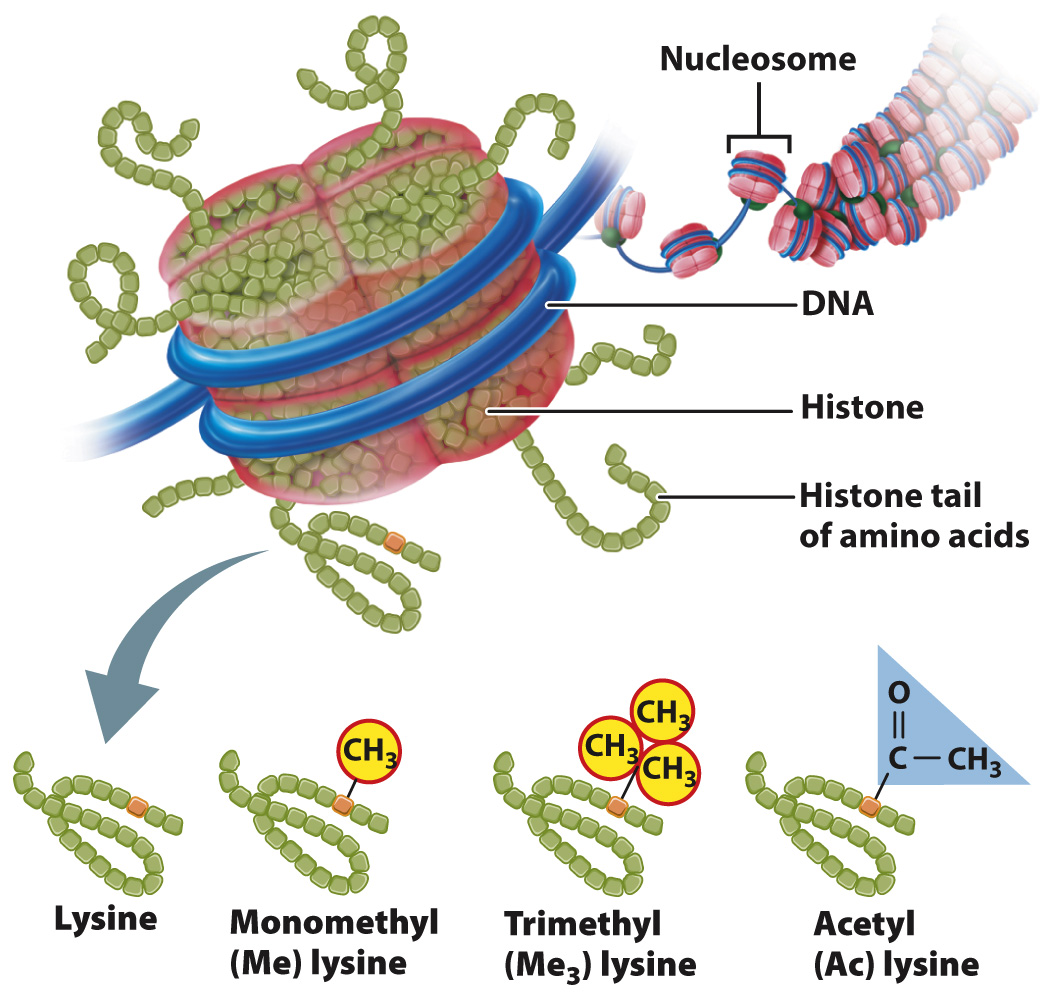
One way in which chromatin is remodeled is by chemical modification of the histones around which DNA is wound (Fig. 19.2). Modification usually occurs on histone tails, strings of amino acids that protrude from the histone proteins in the nucleosome. Individual amino acids in the tails can be modified by the addition (or later removal) of different chemical groups, including methyl groups (—CH3) and acetyl groups (—COCH3). Most often, methylation or acetylation occurs on the lysine residues of the histone tails. Some of these modifications tend to activate transcription and others to repress transcription. The pattern of modifications of the histone tails constitutes a histone code that affects chromatin structure and gene transcription. Modification of histones takes place at key times in development to ensure that the proper genes are turned on or off, as well as in response to environmental cues.
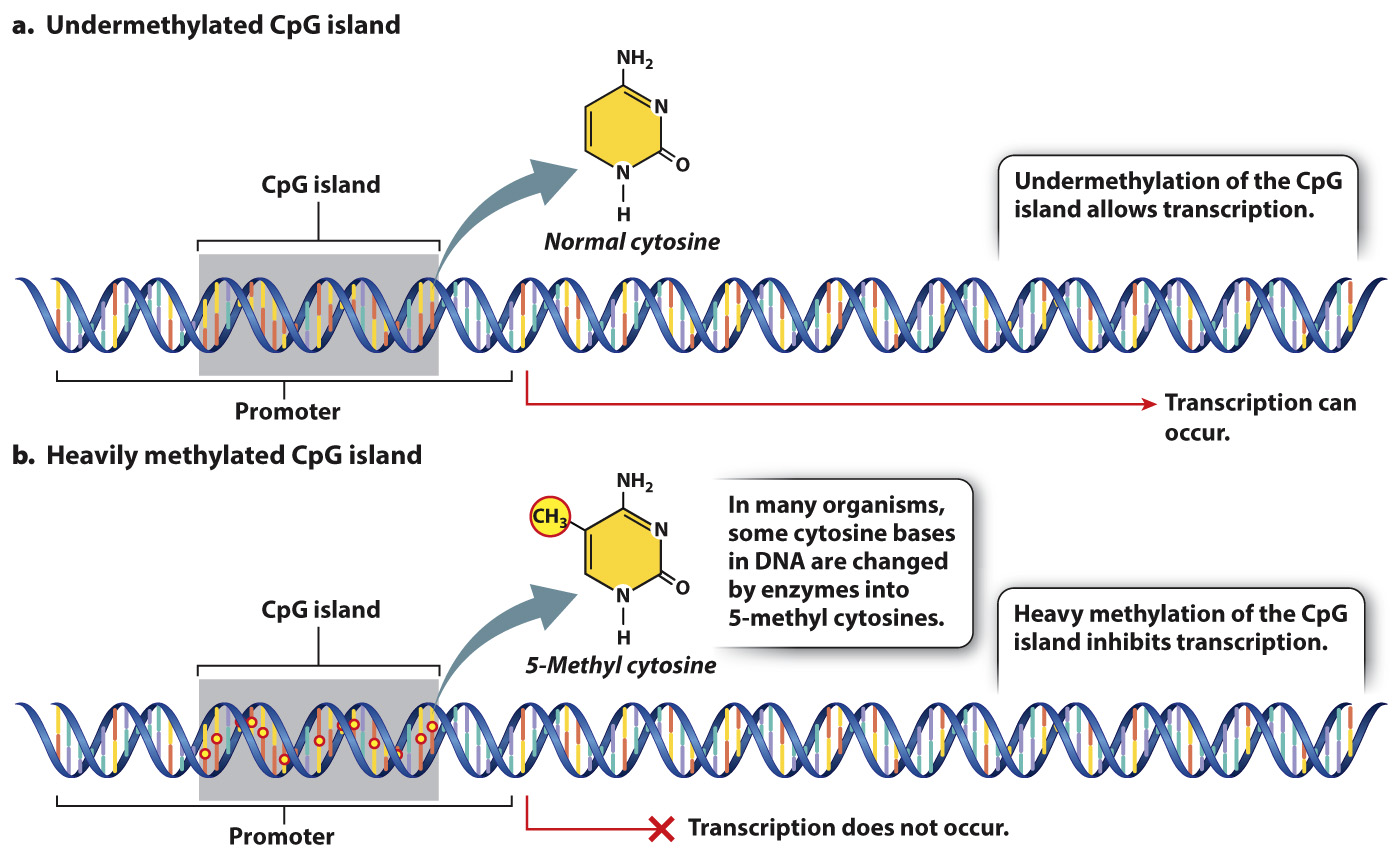
In many eukaryotic organisms, gene expression is also affected by chemical modification of certain bases in the DNA, the most common of which is the addition of a methyl group to the base cytosine (Fig. 19.3). DNA methylation recruits proteins that affect changes in chromatin structure, histone modification, and nucleosome positioning that restrict access of transcription factors to gene promoters. Methylation of cytosines often occurs in cytosine bases that are adjacent to guanosine bases on a DNA strand. Cytosine methylation often occurs in CpG islands, which are clusters of adjacent CG nucleotides located in or near the promoter of a gene. Heavy cytosine methylation is associated with transcriptional repression of the gene near the CpG island. The methylation state of a CpG island can change over time or in response to environmental signals, providing a way to turn genes on or off. Cells sometimes heavily methylate CpG islands of transposable elements or viral DNA sequences that are integrated into the genome, thus preventing the expression of genes in the foreign DNA (Chapter 14).
379
Together, the modification of cytosine bases, changes to histones, and alterations in chromatin structure are often termed epigenetic, from the Greek epi-
In humans and other mammals, about 100 genes are repressed by chemical modification such as DNA methylation in the germ line in a sex-
380
Some of the genes that are imprinted affect the growth rate of the embryo. For example, in matings between lions and tigers that take place in captivity, the offspring of a male tiger with a female lion (a “tigon”) is about the same size as its parents, whereas the offspring of a male lion and a female tiger (a “liger”) is a giant cat—