Translational regulation controls the rate, timing, and location of protein synthesis.
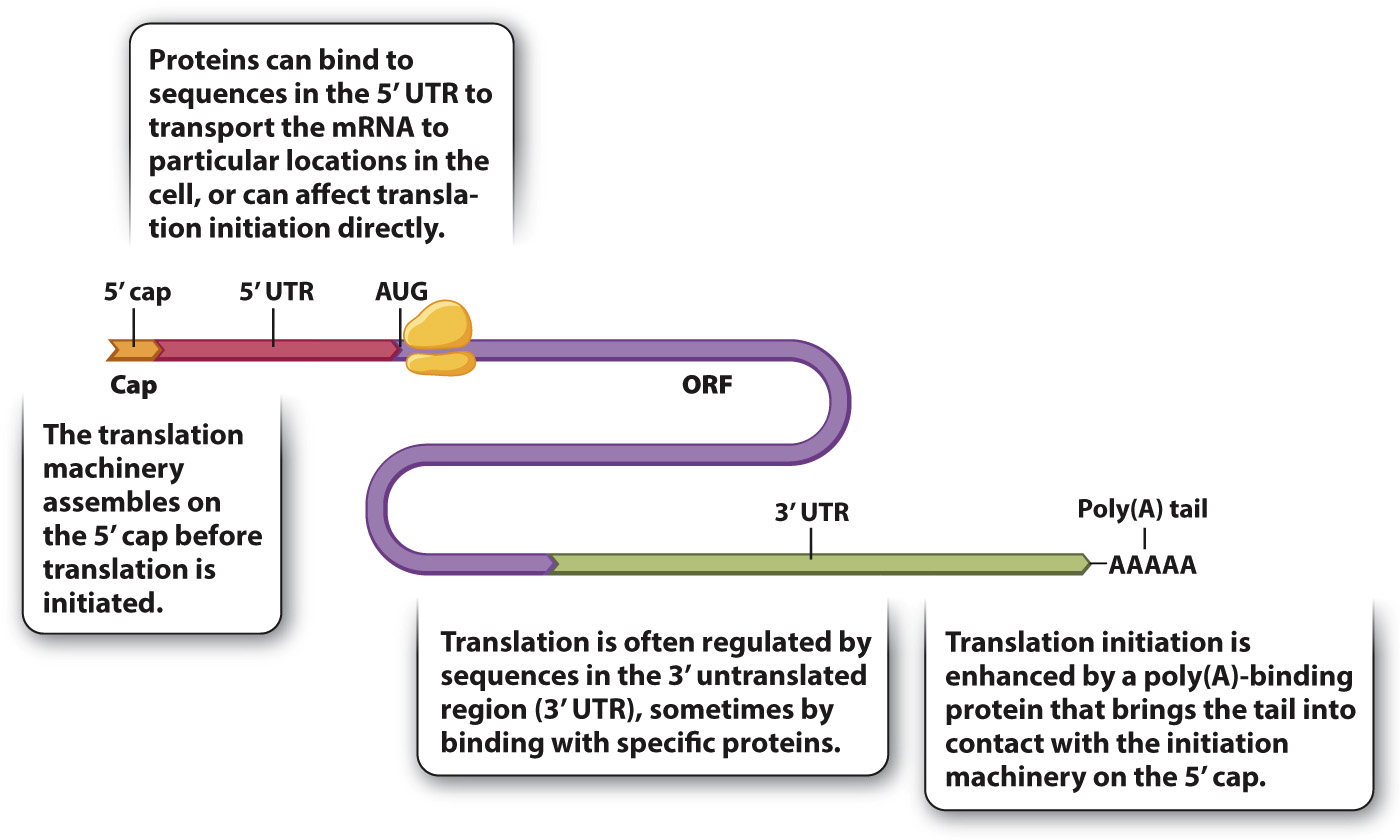
Translation of mRNA into protein provides another level of control of gene expression. Fig. 19.11 shows the structure of a hypothetical mRNA molecule in a eukaryotic cell and highlights some of the features that help regulate its translation (Chapter 4). Not all mRNA molecules have all the features shown, but almost all mRNA molecules have a 5′ cap, a 5′ untranslated region (5′ UTR), an open reading frame (ORF) containing the codons that determine the amino acid sequence of the protein, a 3′ untranslated region (3′ UTR), and a poly(A) tail. The 5′ UTR and 3′ UTR may contain regions that bind with proteins. These RNA-
385
The cap structure is one of the main recognition signals for translation initiation, which requires the coordinated action of about 25 proteins. These proteins are present in most cells in limiting amounts, and so at any one time while some mRNAs from a gene are being translated, other mRNAs transcribed from the same gene may not have a translation initiation complex assembled. Upon formation, the initiation complex moves along the 5′ UTR, scanning for an AUG codon (the initiation codon) to allow the complete ribosome to assemble and begin translation.
The 3′ UTR and the poly(A) tail are also important in translation initiation. The efficiency of translation initiation is greatly increased by physical contact between a protein that binds the poly(A) tail and one that binds the 5′ cap. The physical contact creates a loop in the mRNA, bringing the 3′ end of the mRNA into proximity with the start site for translation. In fact, most mRNA sequences that regulate translation are present in the 3′ UTR.
Although translation initiation is the principal mode of translational regulation, not all mRNA molecules are equally accessible to translation. Among the key variables are the secondary (folded) structure of the 5′ UTR, the distance from the 5′ cap to the AUG initiation codon, and the sequences flanking the AUG initiation codon.