Development proceeds by progressive regionalization and specification.
Nusslein-Volhard and Wieschaus also discovered mutants of the embryo’s developmental genes. They discovered three classes of such mutants, and their analysis showed that genes in the embryo controlling its development are turned on in groups, and that each successive group acts to refine and narrow the pattern of differentiation generated by previous groups. This, too, is a general principle of development in many multicellular organisms.
The anterior–posterior gradient set up by the maternal-effect genes is first narrowed by genes called gap genes (Fig. 20.9), each of which is expressed in a broad region of the embryo. The name “gap gene” derives from the phenotype of mutant embryos, which are missing groups of adjoining segments, leaving a gap in the pattern of segments. Fig. 20.9 shows the expression pattern of the gap gene Krüppel, which is expressed in the middle region of the embryo. Mutants of Krüppel lack some thoracic and abdominal segments when they reach the larval stage. Krüppel is expressed in the pattern shown in Fig. 20.9 because it is under the control of the transcription factor Hunchback, which in this embryo is stained in green. High concentrations of Hunchback repress Krüppel transcription entirely, and low concentrations fail to induce Krüppel transcription. Because Hunchback is present in an anterior–posterior gradient (see Fig. 20.8), the pattern of Hunchback expression means that Krüppel is transcribed only in the middle region of the embryo where Hunchback is present but not too abundant.
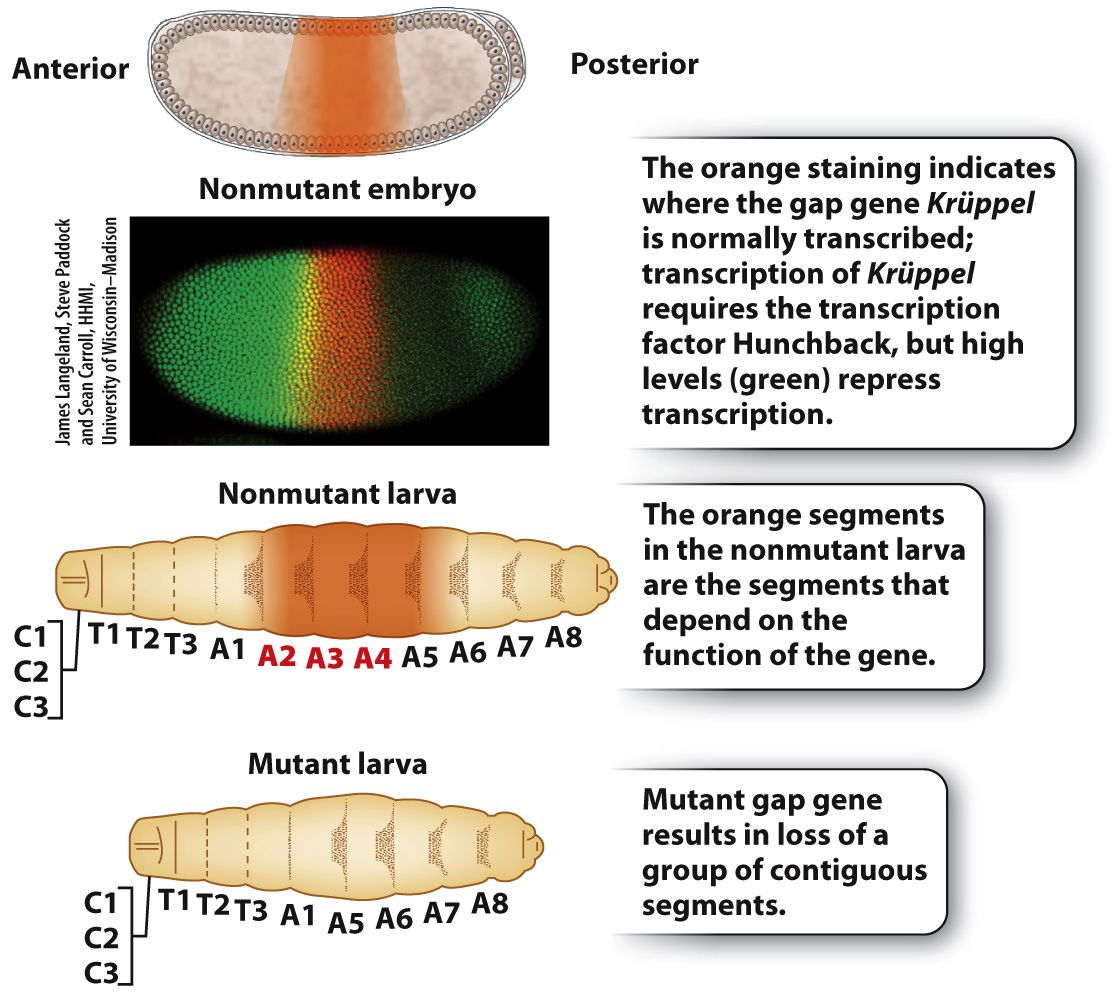
FIG. 20.9 Normal gap-gene expression pattern and mutant phenotype.
The gap genes encode transcription factors that control genes in the next level of the regulatory hierarchy, which consists of pair-rule genes (Fig. 20.10). Pair-rule genes receive their name because larvae with these mutations lack alternate body segments. The example in Fig. 20.10 is hairy, whose mutants lack the odd-numbered thoracic segments and the even-numbered abdominal segments. The pair-rule genes help to establish the uniqueness of each of seven broad stripes across the anterior–posterior axis.
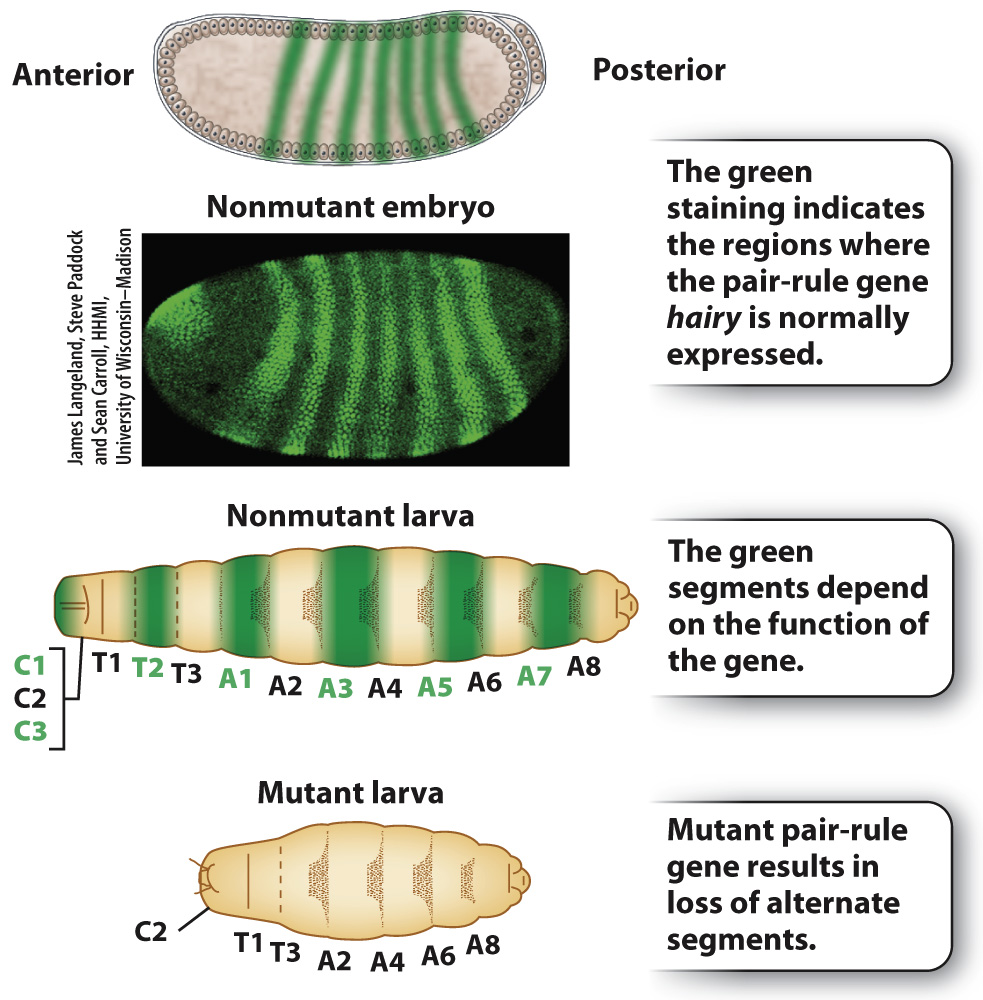
FIG. 20.10 Normal pair-rule gene expression pattern and mutant phenotype.
The pair-rule genes in turn help to regulate the next level in the segmentation hierarchy, which consists of segment-polarity genes (Fig. 20.11). The segment-polarity genes refine the 7-striped pattern still further into a 14-striped pattern. Each of the 14 stripes has distinct anterior and posterior ends determined by the segment-polarity genes. Embryos with mutations in segment-polarity genes lose this anterior–posterior differentiation, with the result that the anterior and the posterior halves of each segment are mirror images. The example in Fig. 20.11 is engrailed, which eliminates the posterior pattern element in each stripe and replaces it with a mirror image of the anterior pattern element.
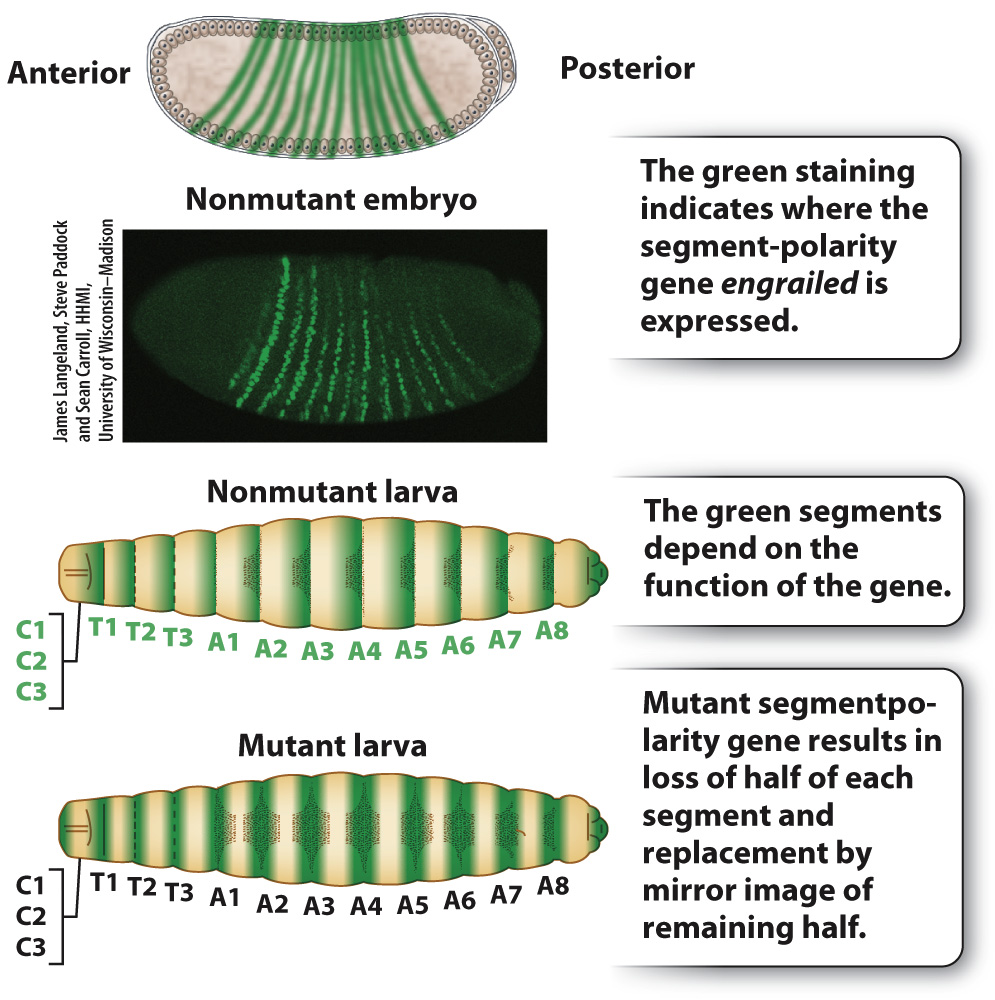
FIG. 20.11 Normal segment-polarity gene expression pattern and mutant phenotype.
Quick Check 4 Would the pattern of segment-polarity gene expression be normal if one or more gap genes were not expressed properly? Why or why not?
Quick Check 4 Answer
The gap genes control the expression of the pair-rule genes, which in turn control the expression of segment-polarity genes. Therefore, if gap-gene expression is not normal, you would predict that segment-polarity gene expression would also not be normal.


