Studies of mitochondrial DNA reveal that modern humans evolved in Africa relatively recently.
For a long time, it was argued that modern humans derive from the Homo ergaster (or H. erectus) populations that spread around the world starting from around the time of the early emigration from Africa 2 million years ago. This idea is called the multiregional hypothesis of human origins because it implies that different Homo ergaster populations throughout Africa and Eurasia evolved in parallel, with some limited gene flow among them, each producing modern H. sapiens populations. In short, racial differences among humans would have evolved over 2 million years in different geographic locations.
This idea was overturned in 1987 by another study from Allan Wilson’s laboratory, which instead suggested that modern humans arose much more recently from Homo ergaster descendants that remained in Africa (sometimes called Homo heidelbergensis; see Fig. 24.6), and are all descended from an African common ancestor dating from around 200,000 years ago. This newer idea is the out-
To test these two hypotheses about human origins, Rebecca Cann, a student in Allan Wilson’s laboratory, analyzed DNA sequences to reconstruct the human family tree. Specifically, she studied sequences of a segment of mitochondrial DNA from people living around the world (Fig. 24.8).
492
HOW DO WE KNOW?
FIG. 24.8
When and where did the most recent common ancestor of all living humans live?
BACKGROUND With the availability of DNA sequence analysis tools, it became possible to investigate human prehistory by comparing the DNA of living people from different populations.
HYPOTHESIS The multiregional hypothesis of human origins suggests that our most recent common ancestor was living at the time Homo ergaster populations first left Africa, about 2 million years ago.
METHOD Rebecca Cann compared mitochondrial DNA (mtDNA) sequences from 147 people from around the world. This approach required a substantial amount of mtDNA from each individual, which she acquired by collecting placentas from women after childbirth. Instead of sequencing the mtDNA, Cann inferred differences among sequences by digesting the mtDNA with different restriction enzymes, each of which cuts DNA at a specific sequence (Chapter 12). If the sequence is present, the enzyme cuts. If any base in the recognition site of the restriction enzyme has changed in an individual, the enzyme does not cut. The resulting fragments were then separated by gel electrophoresis. By using 12 different enzymes, Cann was able to assay a reasonable proportion of all the mtDNA sequence variation present in the sample.
Here, we see a sample dataset for four people and a chimpanzee. There are seven varying restriction sites:
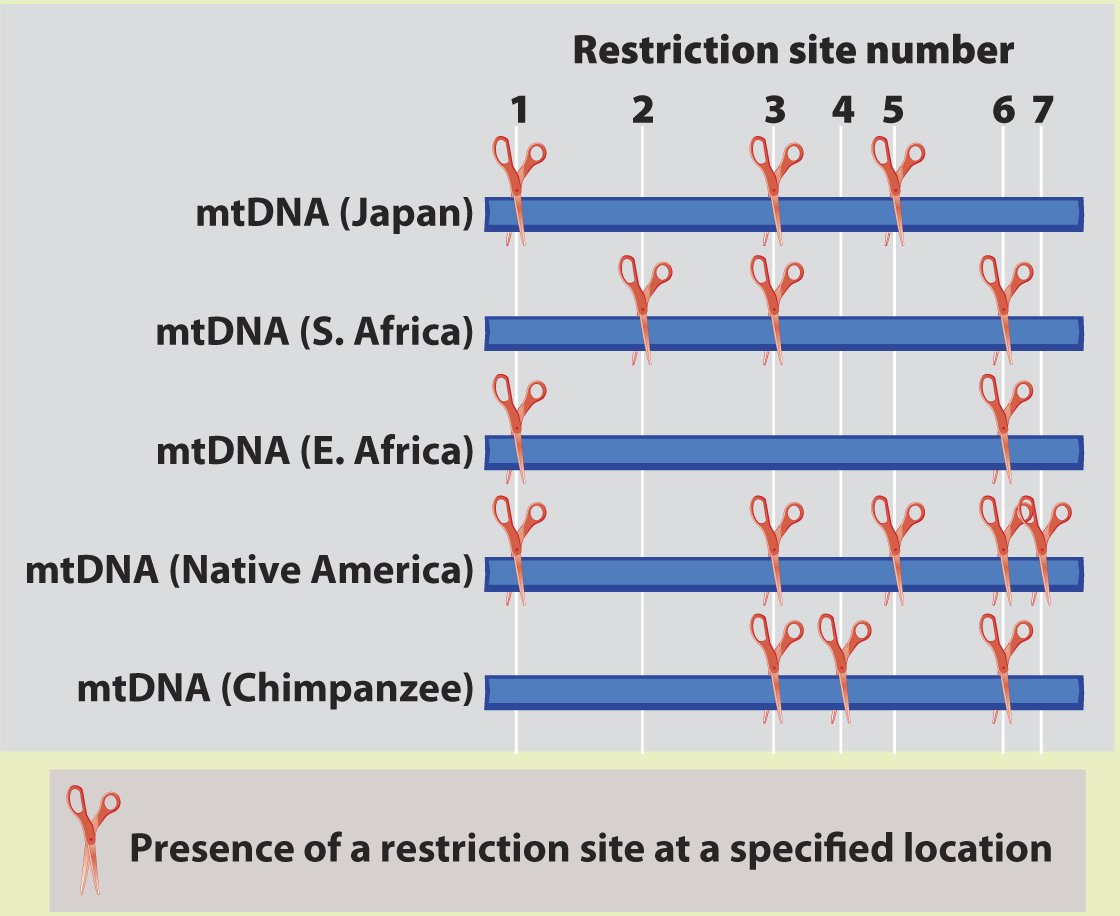
ANALYSIS The data were converted into a family tree by using shared derived characters, described in Chapter 23. By mapping the pattern of changes in this way, the phylogenetic relationships of the mtDNA sequences can be reconstructed. For example, the derived “cut” state at restriction site 1 shared by the East African, Japanese, and Native American sequences implies that the three groups had a common ancestor in which the mutation that created the new restriction site occurred.
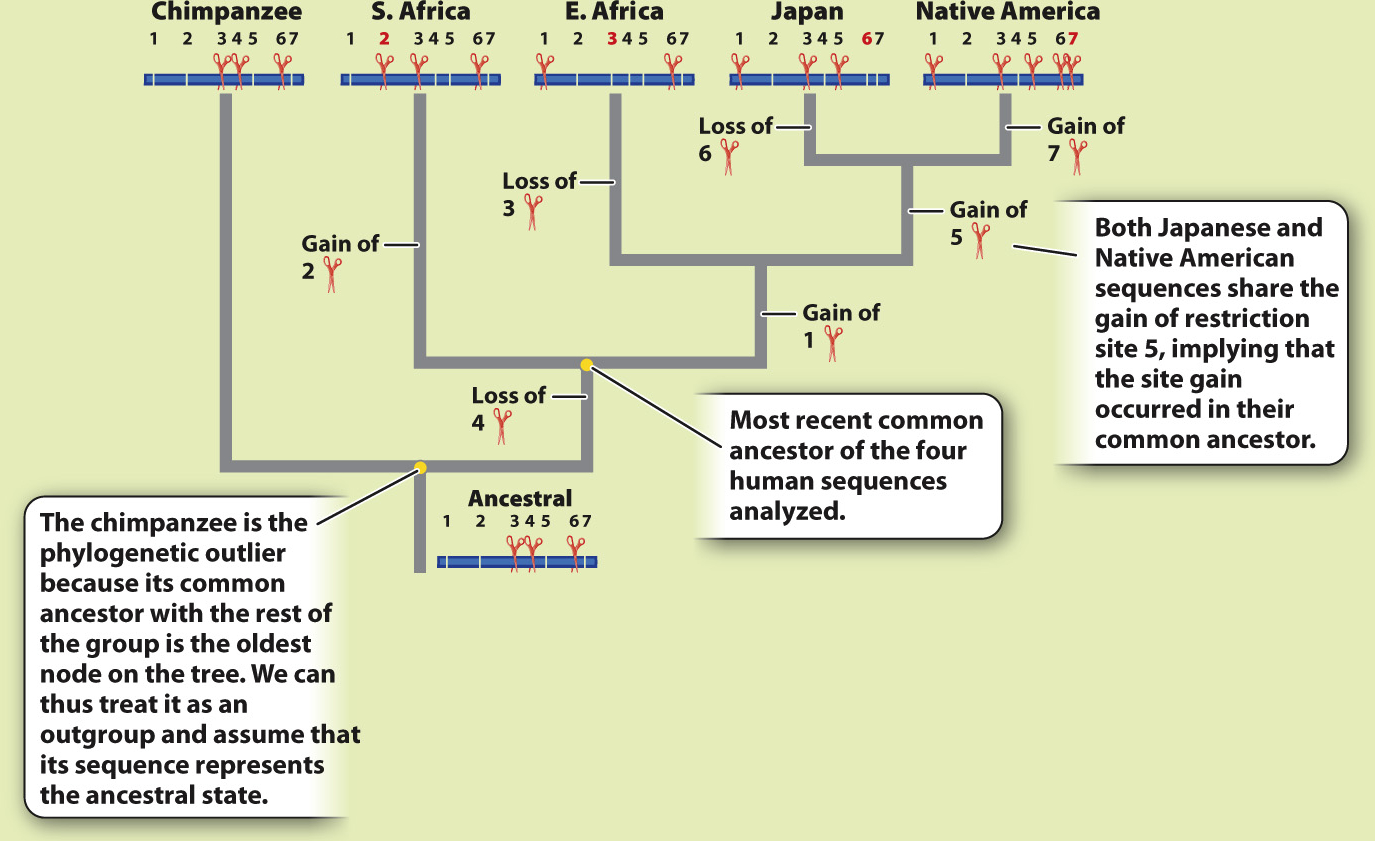
493
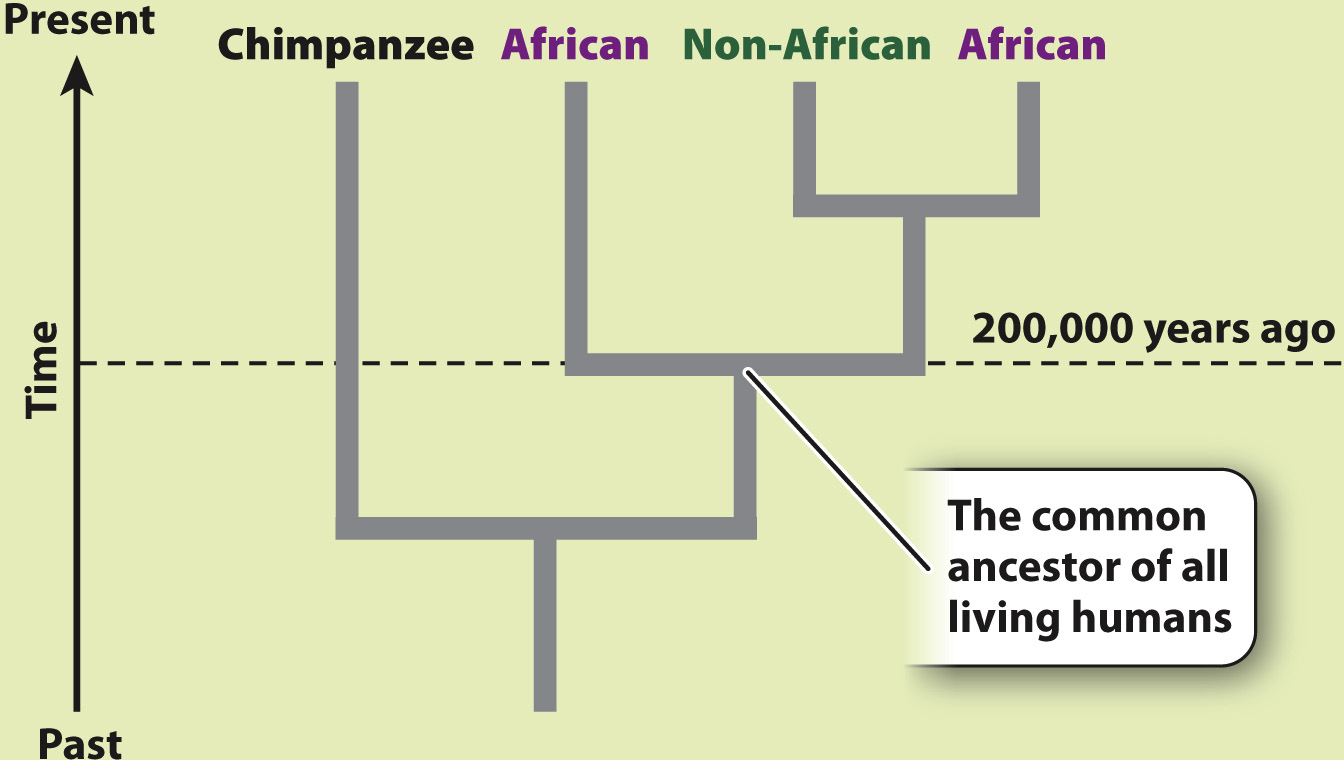
RESULTS AND CONCLUSION A simplified version of Cann’s phylogenetic tree based on mtDNA is shown below. Modern humans arose relatively recently in Africa, and their most common ancestor lived about 200,000 years ago. The multiregional hypothesis is rejected. Also, the data revealed that all non-
FOLLOW-
SOURCE Cann, R. L., M. Stoneking, and A. C. Wilson. 1987. “Mitochondrial DNA and Human Evolution.” Nature 325:31–
Mitochondrial DNA (mtDNA) is a small circle of DNA, about 17,000 base pairs long, found in every mitochondrion (Chapter 17). Cann chose to study mtDNA for several reasons. Although a typical cell contains a single nucleus with just two copies of nuclear DNA, each cell has many mitochondria, each carrying multiple copies of mtDNA, making mtDNA much more abundant than nuclear DNA and therefore easier to extract. More important, however, is its mode of inheritance. All your mtDNA is inherited from your mother in the egg she produces because sperm do not contribute mitochondria to the zygote. This means that there is no opportunity for genetic recombination between different mtDNA molecules, so the only way in which sequence variation can arise is through mutation. In nuclear DNA, by contrast, differences between two sequences can be introduced through both mutation and recombination. Recombination obscures genealogical relationships because it mixes segments of DNA with different evolutionary histories.
Mitochondrial DNA sequences offered Cann a clear advantage. With sequence information from the mtDNA of 147 people from around the world, Cann reconstructed the human family tree (Fig. 24.8). The tree contained two major surprises. First, the two deepest branches—
Second, the tree is remarkably shallow. That is, even the most distantly related modern humans have a relatively recent common ancestor. By calibrating the rate at which mutations occur in mtDNA, Cann was able to estimate the time back to the common ancestor of all modern humans as about 200,000 years. Subsequent analyses have somewhat refined this estimate, but the key message has not changed: The data contradict the multiregional hypothesis, which predicted a number closer to 2 million years.
494
Quick Check 2 What do the multiregional and out-
Quick Check 2 Answer
The multiregional hypothesis suggests that the most recent common ancestor of all living humans lived about 2 million years ago, when H. ergaster first migrated from Africa to colonize Europe and Asia. The out-