Regulatory genes played an important role in the evolution of complex multicellular organisms.
Complex multicellular organisms account for a large proportion of all eukaryotic species. Thus, one evolutionary consequence of complex multicellularity was an increase in biological diversity. Let’s compare the diversity of complex multicellular groups with their simpler phylogenetic sisters. Choanoflagellates, for example, number about 150 species; simple animals such as sponges and jellyfish about 20,000; and complex animals with circulatory systems perhaps as many as 10 million. Plants and their relatives show a comparable pattern. There are about 6000 species of green algae on the branch that includes land plants, but about 400,000 species of anatomically complex land plants capable of bulk flow. Similarly, complex fungi and red algae include more species by an order of magnitude than their simpler relatives.
How did this immense diversity arise? At one level, the answer is functional and ecological. Complex multicellular organisms can perform a range of functions that simpler organisms cannot, and so they have evolved many specific types of interaction with other organisms and the physical environment. Carnivory, for example, requires both sophisticated sensory systems and jaws or limbs to locate and capture prey, but once established, carnivorous animals radiated throughout the oceans and, eventually, on land. This explanation, however, prompts another question: What is the genetic basis for the bewildering range of sizes and shapes displayed by complex multicellular organisms?
The answer has to do with the network of genes that guide development. Since the 1950s, biologists have learned a remarkable amount about this genetic network and its host of interacting genes (Chapter 20). Many of the genes involved in development are what we might consider middle managers: A molecular signal induces the expression of a gene, and the protein product, in turn, prompts the expression or repression of another gene. It is the complex interplay of these genetic switches—
593
HOW DO WE KNOW?
FIG. 28.16
What controls color pattern in butterfly wings?
BACKGROUND Butterfly wings commonly show a striking pattern of color, including circular features known as eyespots. Eyespots are adaptive, for example in deterring predation by birds.
HYPOTHESIS The expression of regulatory genes during wing development governs eyespot formation on wing surfaces.
EXPERIMENT Paul Brakefield and his colleagues mapped the expression of a regulatory gene called Distalless in developing butterfly wings. Distalless genes were fused to genes for green fluorescent protein (GFP), a protein that glows vivid green when illuminated under blue light. GFP is then expressed along with Distalless, allowing the spatial pattern of Distalless expression in developing wings to be visualized.
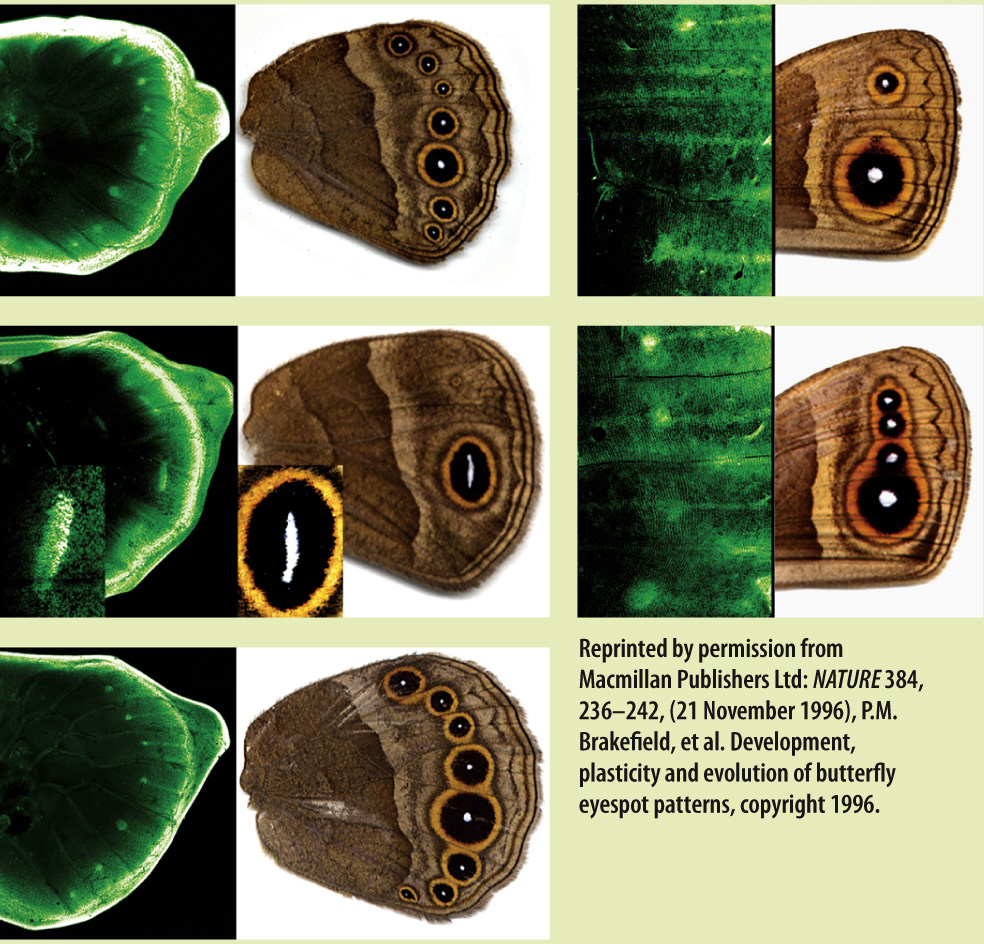
RESULTS The expression pattern of Distalless (on the left in each panel of the figure) in developing wings closely resembles the pattern of eyespots on the wing (on the right in each panel). The two patterns match for both wild-
CONCLUSION Regulatory genes play an important role in butterfly wing coloration, and mutations in these genes can account for differences in wing color patterns among species.
SOURCE Brakefield, P. M., et al. 1996. “Development, Plasticity and Evolution of Butterfly Eyespot Patterns.” Nature 384: 236–
It makes sense that if regulatory genes guide development of the body in each complex multicellular species, then mutations in regulatory genes may account for many of the differences we observe among different species. Our growing understanding of how developmental genes underpin evolutionary change has given rise to a whole new field of biology called evolutionary-
In evo-