Primary transcripts in eukaryotes undergo several types of chemical modification.
In eukaryotes, the nuclear envelope is a barrier between the processes of transcription and translation. Transcription takes place in the nucleus, and translation in the cytoplasm. The separation allows for a complex chemical modification of the primary transcript, known as RNA processing, which converts the primary transcript into the finished mRNA, which can then be translated by the ribosome.
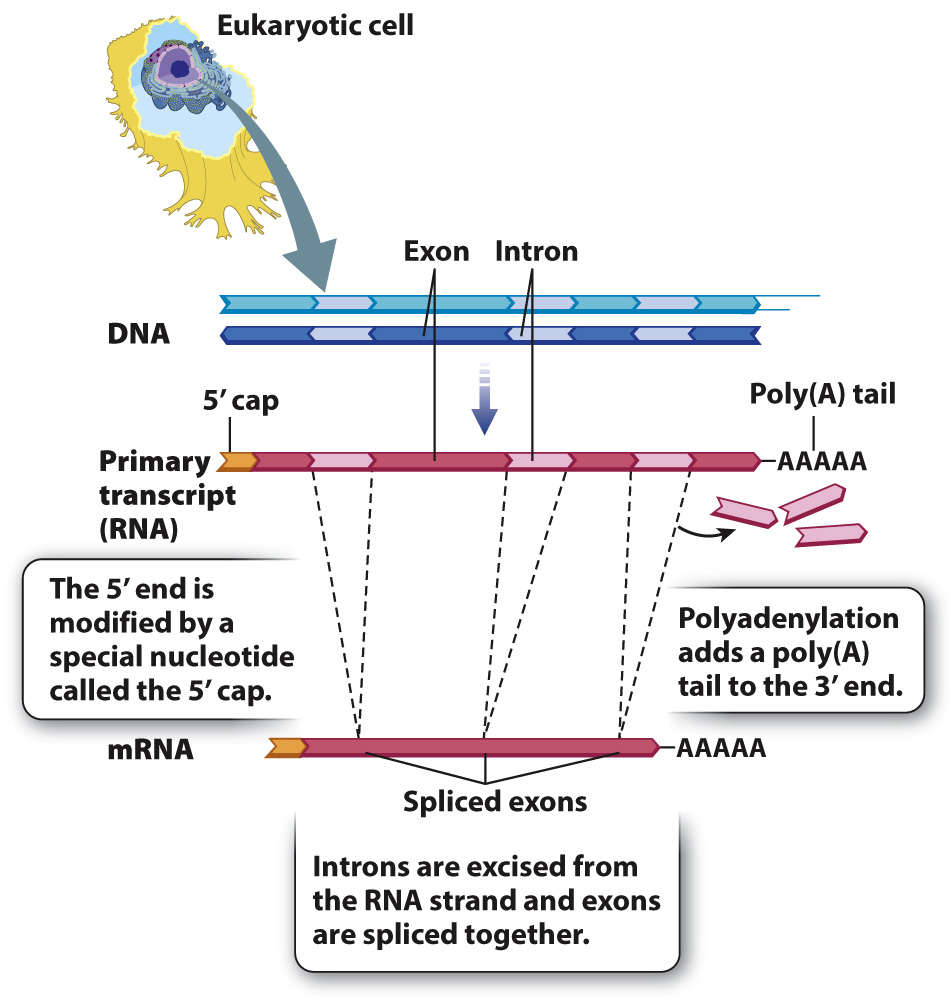
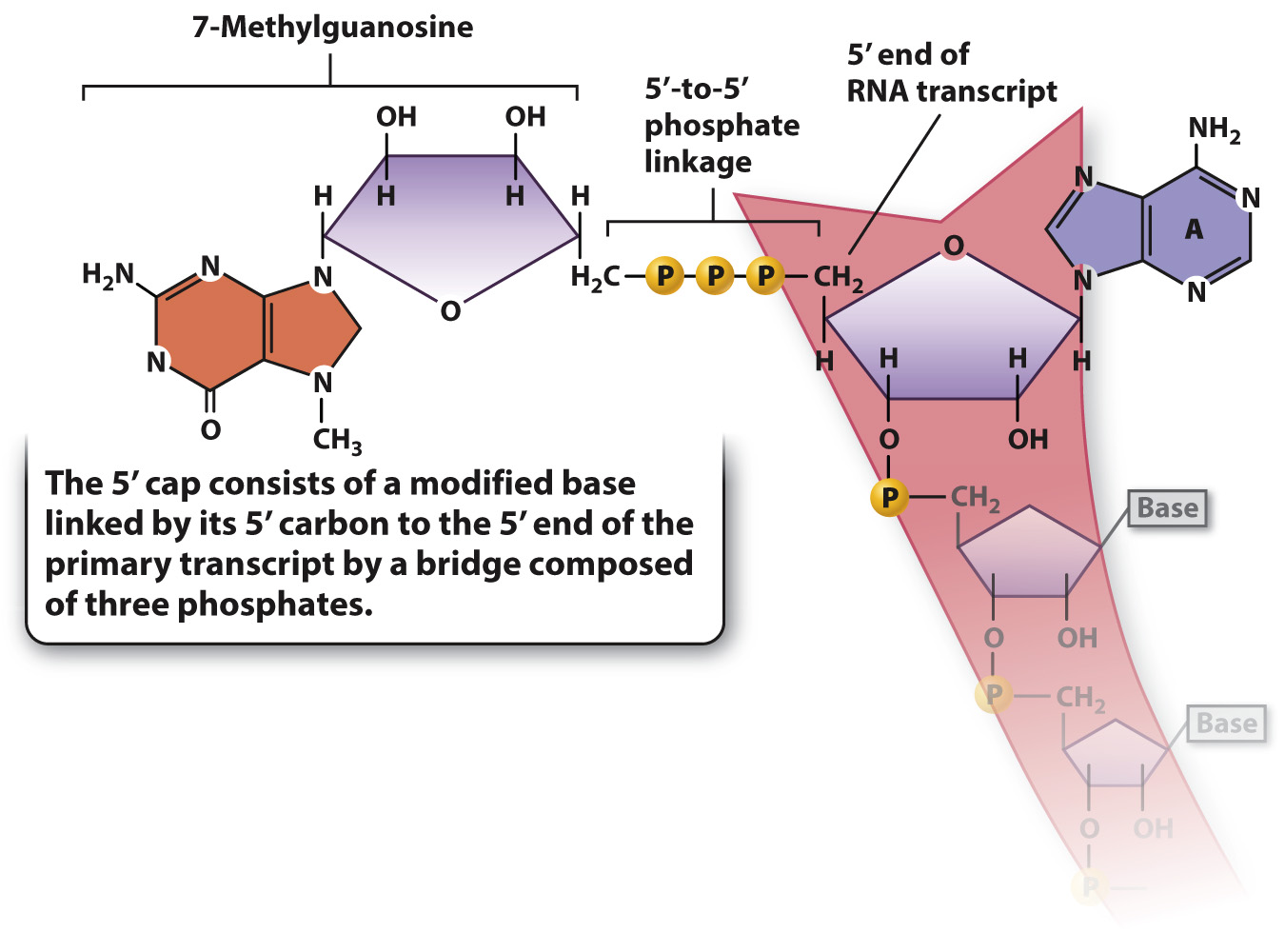
RNA processing consists of three principal types of chemical modification, illustrated in Fig. 3.21. First, the 5′ end of the primary transcript is modified by the addition of a special nucleotide attached in an unusual linkage. This addition is called the 5′ cap, and it consists of a modified nucleotide called 7-
The second major modification of eukaryotic primary transcripts is polyadenylation, the addition of a string of about 250 consecutive A-
Not every stretch of the RNA transcript ends up being translated into protein. Transcripts in eukaryotes often contain regions of protein-
65
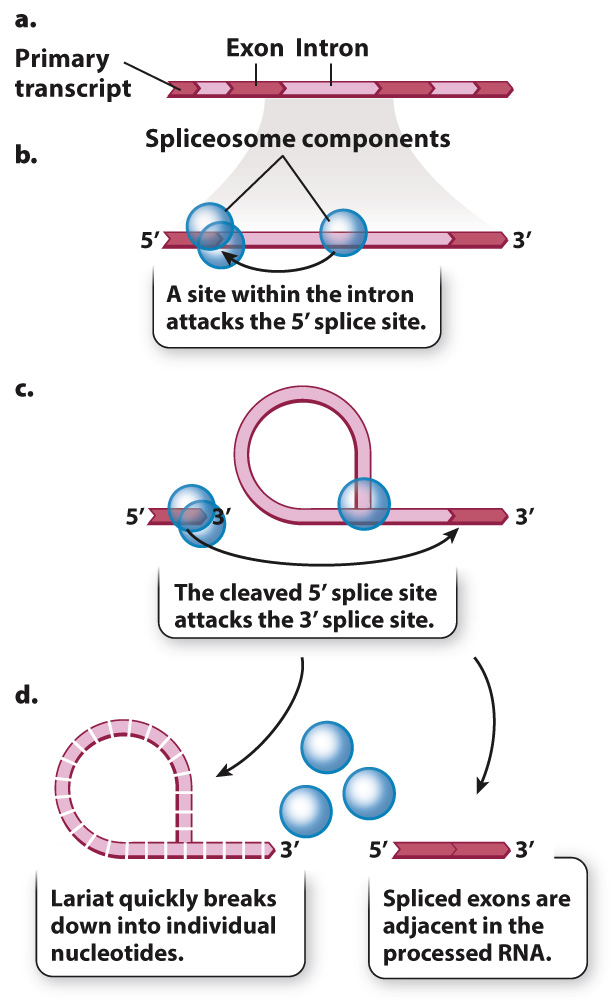
The mechanism of splicing is outlined in Fig. 3.23. In the first step, the spliceosome brings a specific sequence within the intron into proximity with the 5′ end of the intron, at a site known as the 5′ splice site (Fig. 3.23a). The proximity enables a reaction that cuts the RNA at the 5′ splice site, and the cleaved end of the intron connects back on itself forming a loop and tail called a lariat (Fig. 3.23b). In the next step, the spliceosome brings the 5′ splice site close to the splice site at the 3′ end of the intron. The 5′ splice site attacks the 3′ splice site (Fig. 3.23c), cleaving the bond that holds the lariat on the transcript and attaching the ends of the exons to each other. The result is that the exons are connected and the lariat is released (Fig. 3.23d). The lariat making up the intron is quickly broken down into its constituent nucleotides.
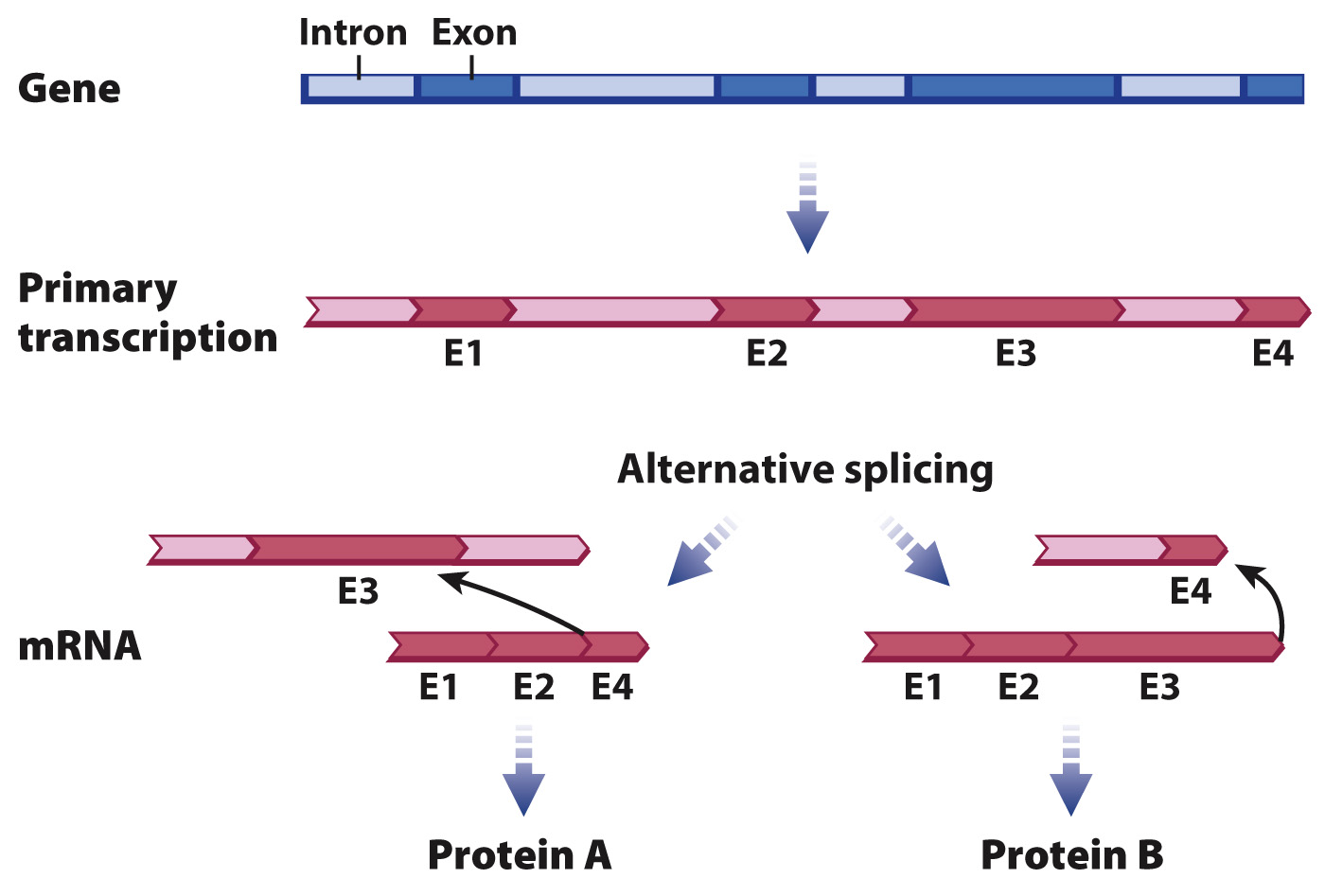
About 90% of all human genes contain at least one intron. Although most genes contain 6 to 9 introns, the largest number is 147, found in a muscle gene. Most introns are just a few thousand nucleotides in length, but about 10% are longer than 10,000 nucleotides. The presence of multiple introns in most genes allows for a process known as alternative splicing, in which primary transcripts from the same gene can be spliced in different ways to yield different mRNAs and therefore different protein products (Fig. 3.24). More than 80% of human genes are alternatively spliced. In most cases, the alternatively spliced forms differ in whether a particular exon is or is not removed from the primary transcript along with its flanking introns. Alternative splicing allows the same transcript to be processed in diverse ways to produce mRNA molecules with different combinations of exons coding for different proteins.
Quick Check 4 When a region of DNA that contains the genetic information for a protein is isolated from a bacterial cell and inserted into a eukaryotic cell in a proper position between a promoter and a terminator, the resulting cell usually produces the correct protein. But when the experiment is done in the reverse direction (eukaryotic DNA into a bacterial cell), the correct protein is often not produced. Can you suggest an explanation?
Quick Check 4 Answer
The eukaryotic DNA sequence contains introns, which the bacterial cell cannot splice out properly, and so the correct protein is not produced from the information in the bacterial RNA transcript.