Amino acids differ in their side chains.
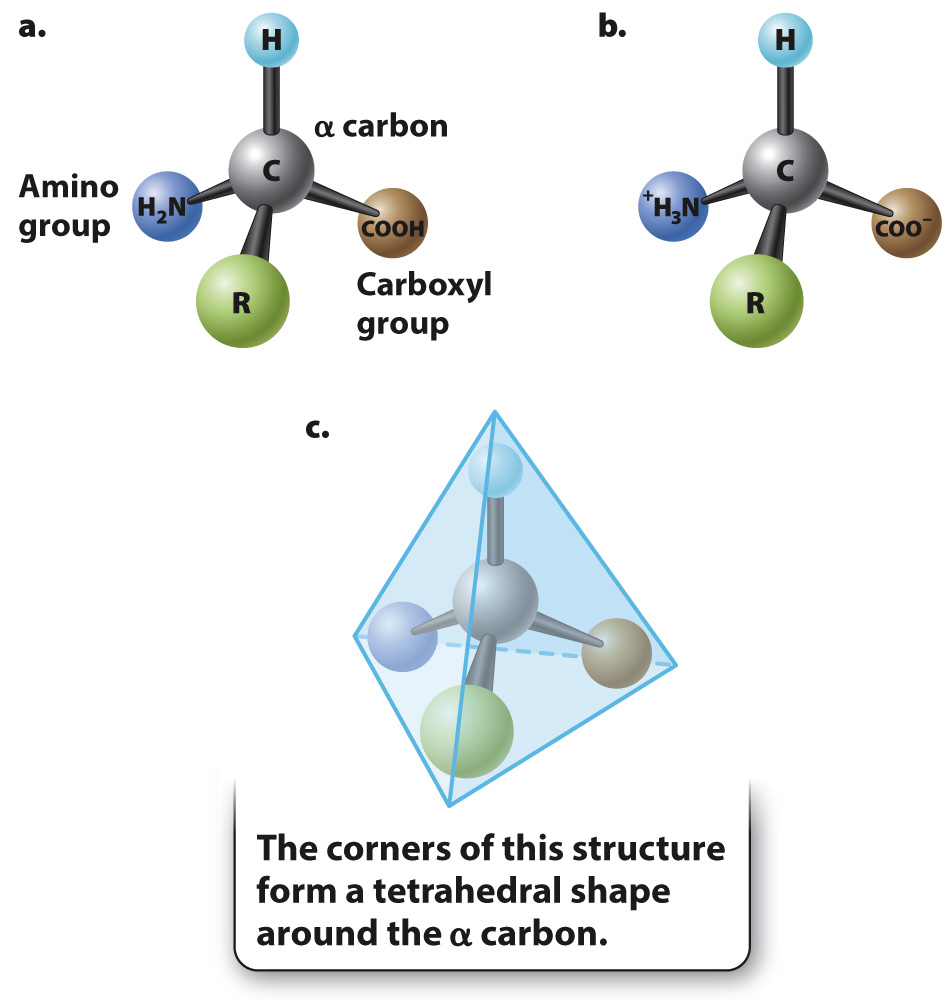
The general structure of an amino acid was discussed in Chapter 2 and is shown again in Fig. 4.1. It consists of a central carbon atom, called the α (alpha) carbon, connected by covalent bonds to four different chemical groups (Fig. 4.1a): an amino group (–NH2, shown in dark blue), a carboxyl group (–COOH, shown in brown), a hydrogen atom (–H, shown in light blue), and a variable side chain or R group (shown in green). In the environment of a cell, where the pH is in the range 7.35–![]() and the carboxyl group loses a proton to become –COO– (Fig. 4.1b). The four covalent bonds from the α carbon are at equal angles. As a result, an amino acid forms a tetrahedron, a pyramid with four triangular faces (Fig. 4.1c).
and the carboxyl group loses a proton to become –COO– (Fig. 4.1b). The four covalent bonds from the α carbon are at equal angles. As a result, an amino acid forms a tetrahedron, a pyramid with four triangular faces (Fig. 4.1c).
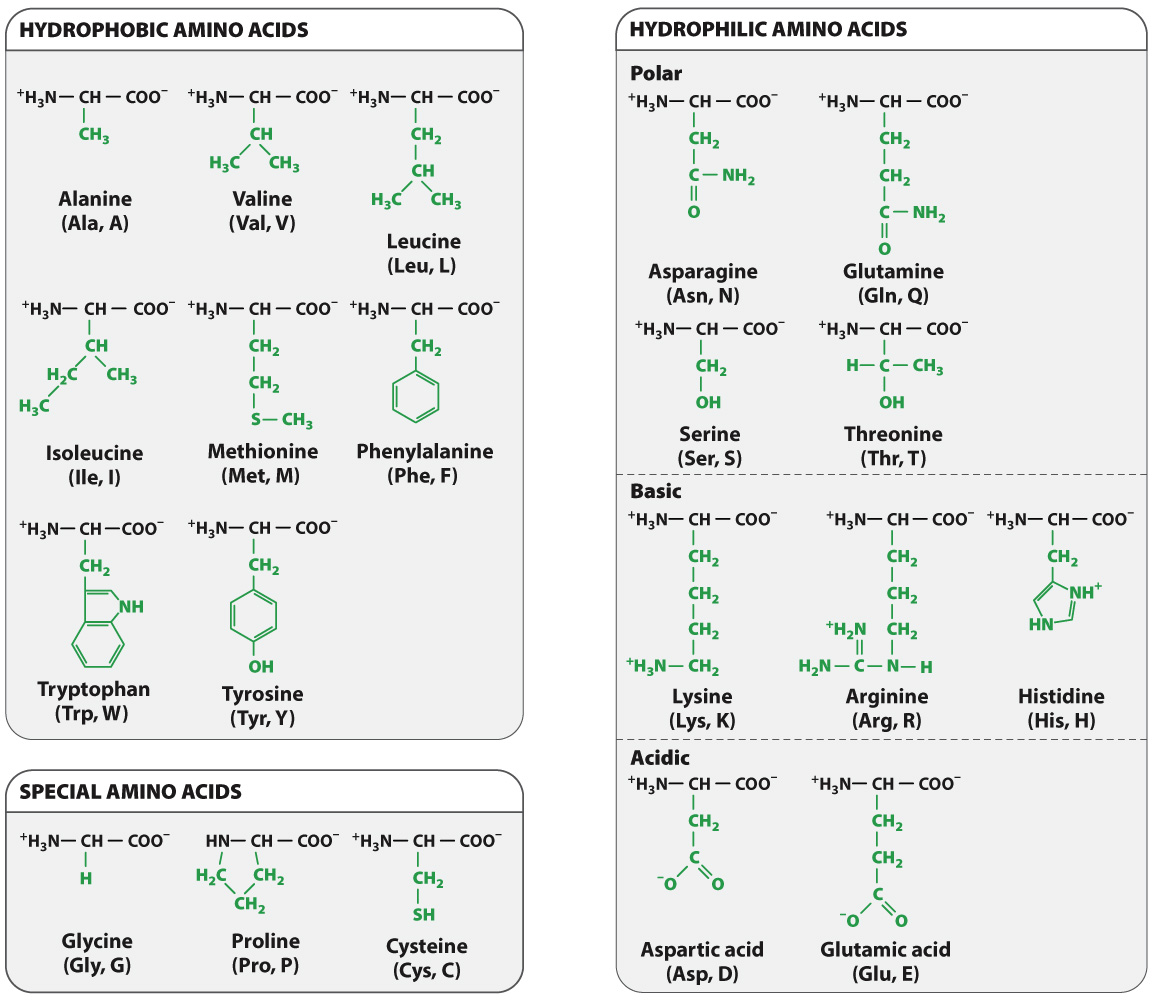
The R groups of the amino acids differ from one amino acid to the next. They are what make the “letters” of the amino acid “alphabet” distinct from one another. Just as letters differ in their shapes and sounds—
The chemical structures of the 20 amino acids commonly found in proteins are shown in Fig. 4.2. The R groups (shown in green) are chemically diverse and are grouped according to their properties, with a particular emphasis on whether they are hydrophobic or hydrophilic, or have special characteristics that might affect a protein’s structure. These properties strongly influence how a polypeptide folds, and hence the three-
71
Hydrophobic amino acids are those that do not readily interact with water or form hydrogen bonds. Most hydrophobic amino acids have nonpolar R groups composed of hydrocarbon chains or uncharged carbon rings. Because water molecules in the cell form hydrogen bonds with each other instead of with the hydrophobic R groups, the hydrophobic R groups tend to aggregate with each other. Their aggregation is also stabilized by weak van der Waals forces (Chapter 2), in which asymmetries in electron distribution create temporary charges in the interacting molecules, which are then attracted to each other. The tendency for hydrophilic water molecules to interact with each other and for hydrophobic molecules to interact with each other is the very same tendency that leads to the formation of oil droplets in water. This is also the reason why most hydrophobic amino acids tend to be buried in the interior of folded proteins, where they are kept away from water.
Amino acids with polar R groups have a permanent charge separation, in which one end of the R group is slightly more negatively charged than the other. As we saw in Chapter 2, polar molecules are hydrophilic, and they tend to form hydrogen bonds with each other or with water molecules.
72
The R groups of the basic and acidic amino acids are strongly polar. At the pH of a cell, the R groups of the basic amino acids gain a proton and become positively charged, whereas those of the acidic amino acids lose a proton and become negatively charged. Because the R groups of these amino acids are charged, they are usually located on the outside surface of the folded molecule. The charged groups can also form ionic bonds with each other and with other charged molecules in the environment, in which a negatively charged group or molecule bonds with a positively charged group or molecule. This ability to bind another molecule of opposite charge is one important way in which proteins can associate with each other or with other macromolecules such as DNA.
The properties of several amino acids are noteworthy because of their effect on protein structure. These amino acids include glycine, proline, and cysteine. Glycine is different from the other amino acids because its R group is hydrogen, exactly like the hydrogen on the other side of the α carbon, and therefore it is not asymmetric. All of the other amino acids have four different groups attached to the α carbon and are asymmetric. In addition, glycine is nonpolar and small enough to tuck into spaces where other R groups would not fit. The small size of glycine’s R group also allows for freer rotation around the C–
Proline is also distinctive, but for a different reason. Note how its R group is linked back to the amino group. This linkage creates a kink or bend in the polypeptide chain and restricts rotation of the C–
Cysteine makes a special contribution to protein folding through its –SH group. When two cysteine side chains in the same or different polypeptides come into proximity, they can react to form an S–