5.18: DNA is an individual identifier: the uses and abuses of DNA fingerprinting.
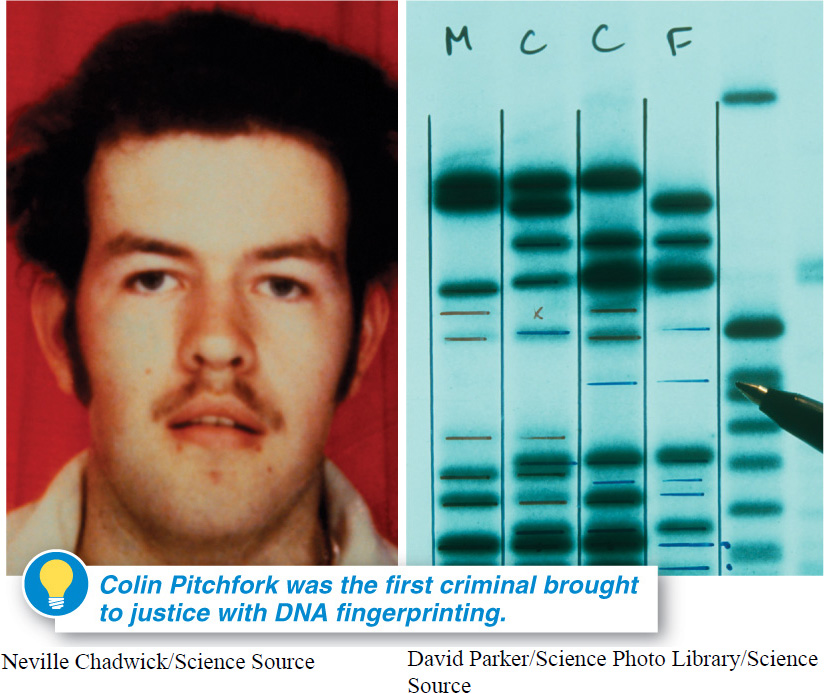
In another time, Colin Pitchfork, a murderer and rapist, would have walked free. But in 1987, he was captured and convicted, betrayed by his DNA, and is now serving two life sentences in prison. Pitchfork’s downfall began when he raped and murdered two 15-
At the time, British biologist Alec Jeffreys made the important discovery that there are small pieces of DNA in human chromosomes that are tremendously variable in their base sequences. In much the same way that each person has a driver’s license number or social security number that differs from everyone else’s, these DNA fragments are so variable that it is extremely unlikely that two people would have identical sequences at these locations. Thus, a comparison between these regions in a person’s DNA sample and in DNA-
Jeffreys analyzed DNA left by the murderer-
221
DNA fingerprinting is now used extensively in forensic investigations, in much the same way that regular fingerprints have been used for the past 100 years. But regular fingerprinting is limited in its usefulness for many crimes because, often, no fingerprints are left behind. DNA samples are frequently left behind, though, usually in the form of semen, blood, hair, skin, or other tissue. As a consequence, this technology has been directly responsible for bringing thousands of criminals to justice and, as we saw at the beginning of the chapter, for establishing the innocence of more than 300 people wrongly convicted of murder and other capital crimes. Let’s examine how DNA fingerprinting is done, why it is such a powerful forensic tool, and why it is not foolproof.
The DNA from different humans is almost completely identical. More than 99.9% of the DNA sequences of two individuals are the same, because we’re all of the same species and thus share a common evolutionary history. Even so, in comparing two individuals’ genomes of three billion base pairs each, a one-
Among these thousands of variable regions, one particular type is used for the determination of a person’s genetic fingerprint. These regions are called STRs (for short tandem repeats) and are characterized by a short sequence (commonly four or five nucleotides) that repeats over and over within the region. The number of repeats is what varies among individuals.
An individual—
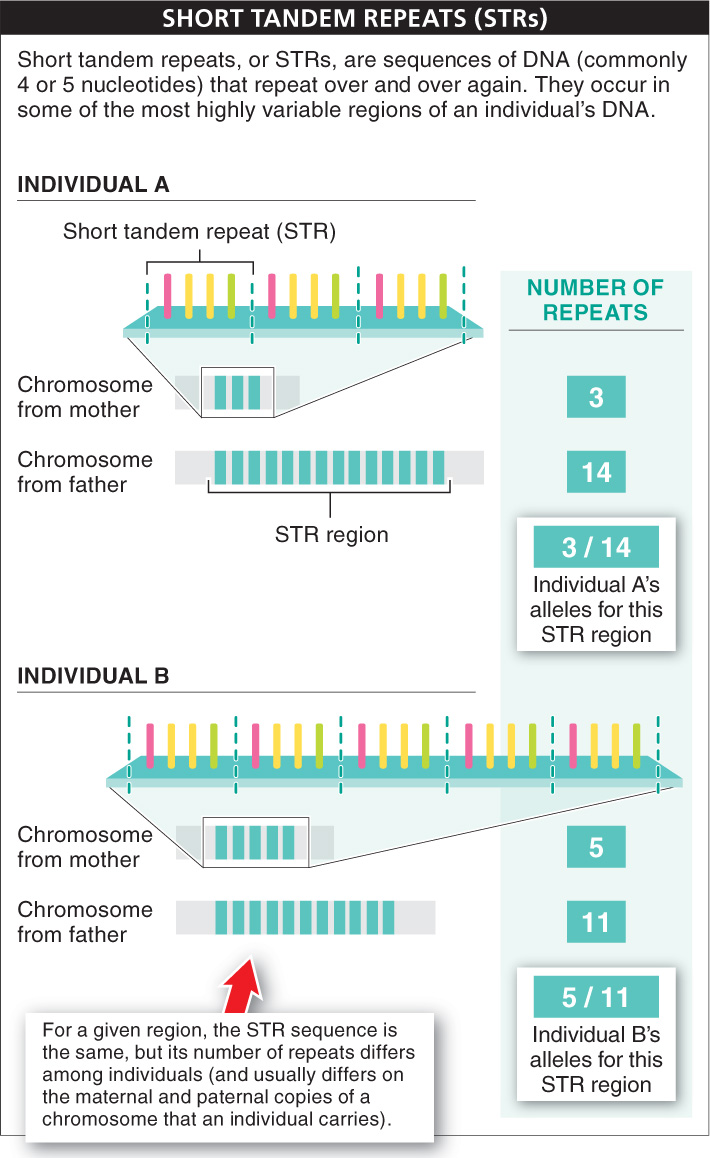
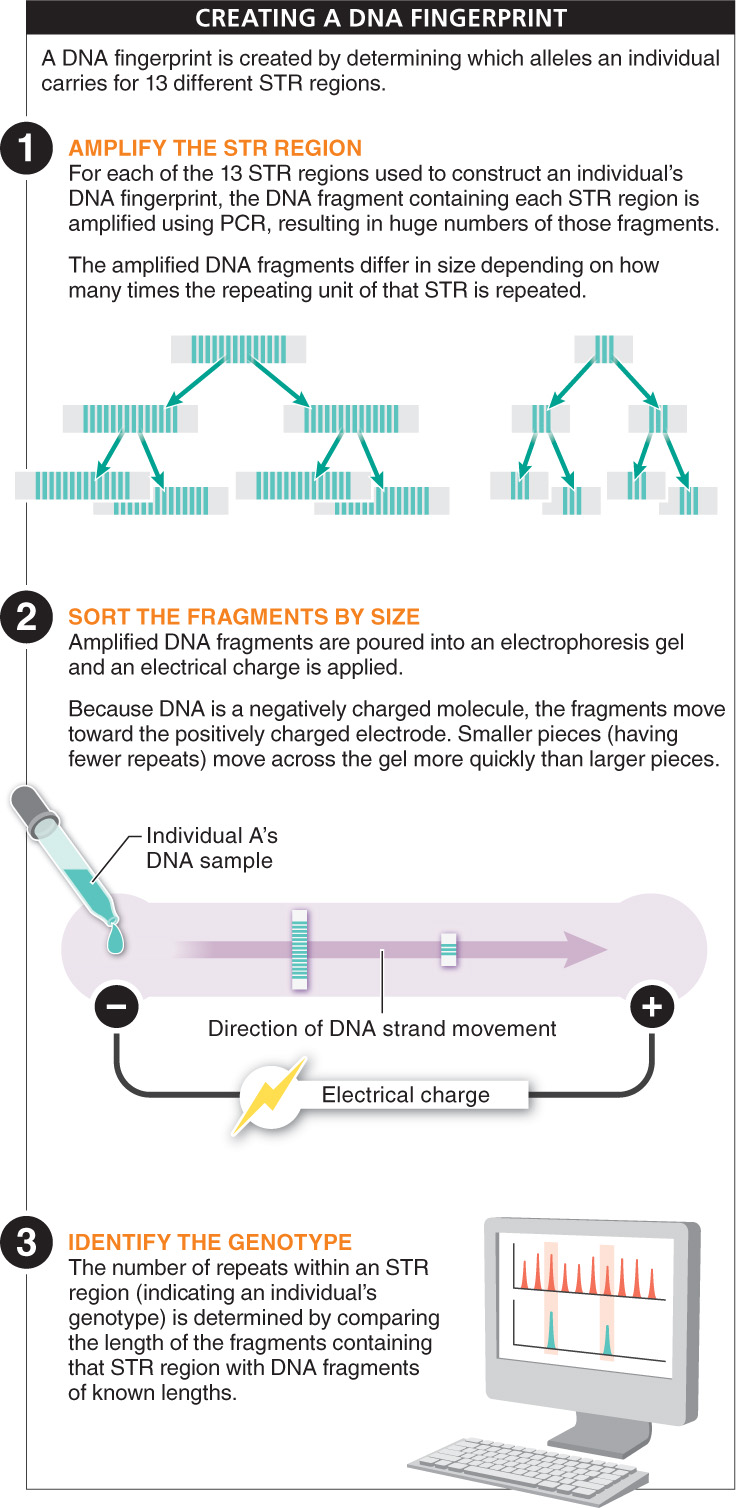
For an STR region within the human genome, there typically are about 10 different alleles that occur within a population, and each is shared by about 10% of all individuals in a population. So the likelihood that two individuals carry the same two alleles is about 1 in 100. This is unlikely, but given enough people, many are likely to carry the same alleles. If two different STR regions are analyzed, the likelihood that two individuals have the same four alleles is 1/10 × 1/10 × 1/10 × 1/10, or 1/10,000. This is much rarer, but still would lead to multiple individuals within the same large city carrying the same four alleles. The real power of DNA fingerprinting comes from simultaneously determining the alleles an individual carries (that is, their genotype) not simply at one or two STR locations but at 13 different STR locations. This is the number used by the FBI in constructing DNA fingerprints in the United States, and it makes the probability that two individuals would have exactly the same genotype extremely low (see Figure 5-
222
For each STR locus analyzed, an individual’s genotype is determined by using PCR to amplify that region, then measuring the length of the STR region using a technique called electrophoresis, which allows researchers to see how long a stretch of DNA is. The length of the region can then be used to determine the number of times the STR is repeated. For a single STR region, an individual’s genotype is expressed by two numbers, reflecting the number of STR repeats in the copies inherited from the mother and from the father. And a person’s full DNA fingerprint is a string of 26 numbers that includes the two numbers for each of 13 STRs.
Q
Question 5.14
What is a DNA fingerprint?
223
In court, a suspect’s genotype might be compared with the DNA fingerprint obtained from evidence found at the crime scene. DNA samples from different people will produce different 26-
In the end, despite universally accepted methods, DNA fingerprinting still is not foolproof. Numerous incidences of human error—
TAKE-HOME MESSAGE 5.18
Comparisons of highly variable DNA regions can be used to identify tissue specimens and determine the individual from whom they came.
What is an STR, and how is the analysis of STRs useful?
224