11.2 Eukaryotic Chromosomes Possess Centromeres and Telomeres
Chromosomes segregate in mitosis and meiosis and remain stable over many cell divisions. These properties of chromosomes arise, in part, from special structural features of chromosomes, including centromeres and telomeres.
Centromere Structure
The centromere is a constricted region of the chromosome to which spindle fibers attach and is essential for proper chromosome movement in mitosis and meiosis (see Chapter 2). The essential role of the centromere in chromosome movement was recognized by early geneticists, who observed the consequences of chromosome breakage. When a chromosome break produces two fragments, one with a centromere and one without (Figure 11.9), the chromosome fragment containing the centromere attaches to spindle fibers and moves to the spindle pole. The fragment lacking a centromere fails to connect to a spindle fiber and is usually lost because it fails to move into the nucleus of a daughter cell during mitosis (see Figure 11.9).
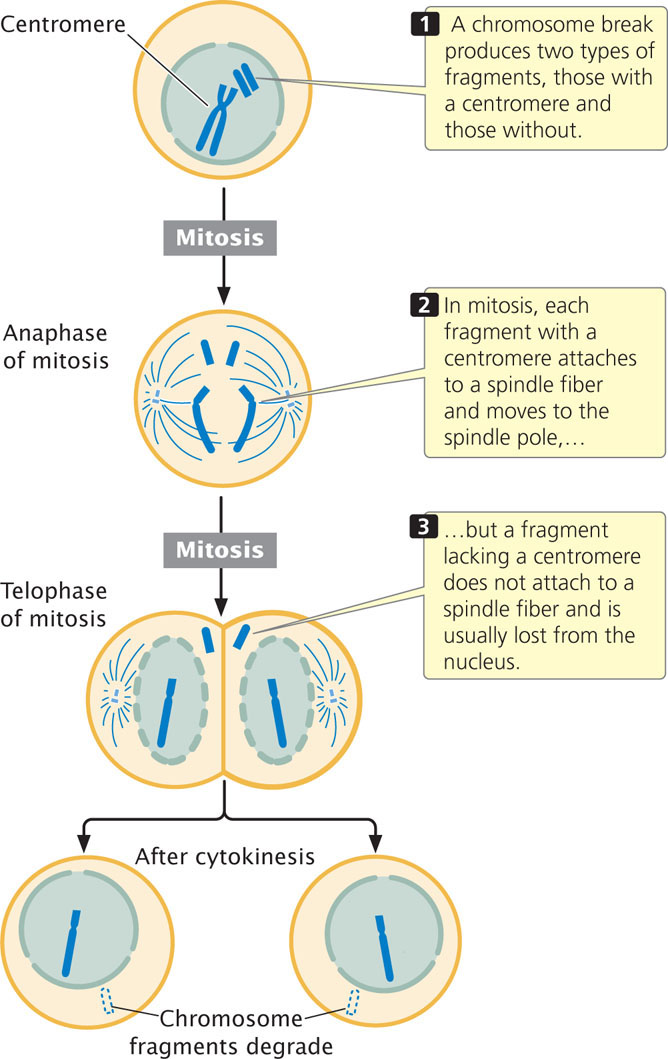
Centromeres are the binding sites for the kinetochore, to which spindle fibers attach. In Drosophila, Arabidopsis, and humans, centromeres span hundreds of thousands of base pairs. Most of the centromere is made up of heterochromatin. Surprisingly, there are no specific sequences that are found in all centromeres, which raises the question of what exactly determines where the centromere is. Research suggests that most centromeres are not defined by DNA sequence but rather by epigenetic changes in chromatin structure. Nucleosomes in the centromeres of most eukaryotes have a variant histone protein called CenH3, which takes the place of the usual H3 histone. The CenH3 histone brings about a change in the nucleosome and chromatin structure, which is believed to promote the formation of the kinetochore and the attachment of spindle fibers to the chromosome.
CONCEPTS
The centromere is a region of the chromosome to which spindle fibers attach. Centromeres display considerable variation in structure and are distinguished by epigenetic alterations to chromatin structure, including the use of a variant H3 histone in the nucleosome.
 CONCEPT CHECK 5
CONCEPT CHECK 5
What happens to a chromosome that loses its centromere?
307
Telomere Structure
Telomeres are the natural ends of a chromosome (see Figure 2.7 and the introduction to this chapter). Pioneering work by Hermann Muller (on fruit flies) and Barbara McClintock (on corn) showed that chromosome breaks produce unstable ends that have a tendency to stick together and enable the chromosome to be degraded. Because attachment and degradation do not happen to the ends of a chromosome that has telomeres, each telomere must serve as a cap that stabilizes the chromosome. Telomeres also provide a means of replicating the ends of the chromosome, which will be discussed in Chapter 12. In 2009, Elizabeth Blackburn, Carol Greider, and Jack Szostak were awarded the Nobel Prize in physiology or medicine for discovering the structure of telomeres and how they are replicated (discussed in Chapter 12).
Telomeres have now been isolated from protozoans, plants, humans, and other organisms; most are similar in structure (Table 11.2). These telomeric sequences usually consist of repeated units of a series of adenine or thymine nucleotides followed by several guanine nucleotides, taking the form 5′-(A or T)mGn-3′, where m is from 1 to 4 and n is 2 or more. For example, the repeating unit in human telomeres is 5′-TTAGGG-3′, which may be repeated from hundreds to thousands of times. The sequence is always oriented with the string of Gs and Cs toward the end of the chromosome, as shown here:
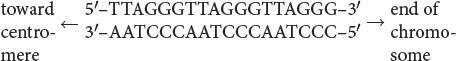
| Organism | Sequence | |
| Tetrahymena (protozoan) | 5′–T T GGGG–3′ 3′–AAC C C C–5′ |
|
| Saccharomyces (yeast) | 5′–T1-6G TG2-3–3′ 3′–A1-6CAC2-3–5′ |
|
| Caenorhabditis (nematode) | 5′–T TAGGC–3′ 3′–AATC C G–5′ |
|
| Vertebrate | 5′–T TAGGG–3′ 3′–AAT CC C–5′ |
|
| Arabidopsis (plant) | 5′–T T TAGGG–3′ 3′–AAAT CCC–5′ |
|
| Source: V. A. Zakian, Science 270:1602, 1995. | ||
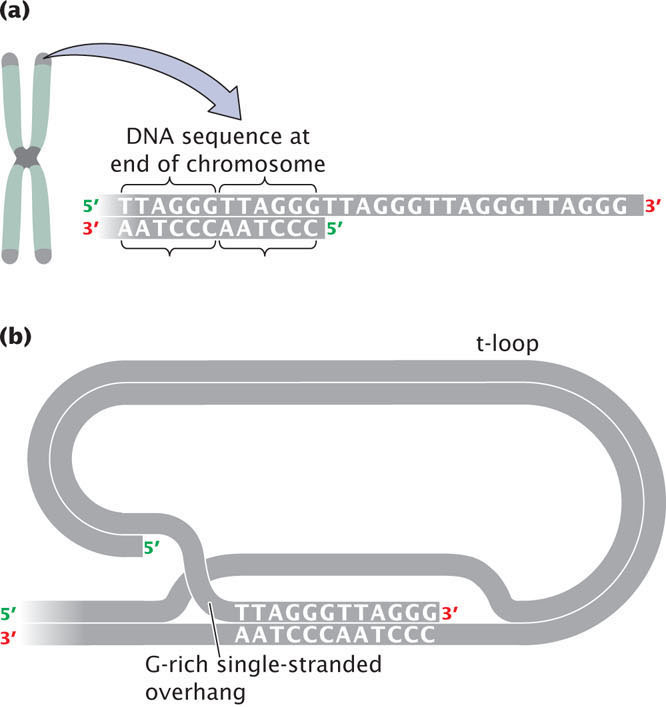
The G-rich strand often protrudes beyond the complementary C-rich strand at the end of the chromosome (Figure 11.10a) and is called the 3′ overhang. The 3′ overhang in the telomeres of mammals is from 50 to 500 nucleotides long. Special proteins bind to the G-rich single-stranded sequence, protecting the telomere from degradation and preventing the ends of chromosomes from sticking together. A multiprotein complex called shelterin binds to telomeres and protects the ends of the DNA from being inadvertently repaired as a double-stranded break in the DNA. In some cells, the single-stranded overhang may fold over and pair with a short stretch of DNA to form a structure called a t-loop, which also functions in protecting the end of the telomere from degradation (Figure 11.10b).
308
CONCEPTS
A telomere is the stabilizing end of a chromosome. At the end of each telomere are many short telomeric sequences.
 CONCEPT CHECK 6
CONCEPT CHECK 6
Which is a characteristic of DNA sequences at the telomeres?
- One strand consists of guanine and adenine (or thymine) nucleotides.
- They consist of repeated sequences.
- One strand protrudes beyond the other, creating some single-stranded DNA at the end.
- All of the above.