12.2 All DNA Replication Takes Place in a Semiconservative Manner
From the three-dimensional structure of DNA proposed by Watson and Crick in 1953 (see Figure 10.7), several important genetic implications were immediately apparent. The complementary nature of the two nucleotide strands in a DNA molecule suggested that, during replication, each strand can serve as a template for the synthesis of a new strand. The specificity of base pairing (adenine with thymine; guanine with cytosine) implied that only one sequence of bases can be specified by each template, and so the two DNA molecules built on the pair of templates will be identical with the original. This process is called semiconservative replication because each of the original nucleotide strands remains intact (conserved), despite no longer being combined in the same molecule; the original DNA molecule is half (semi) conserved during replication.
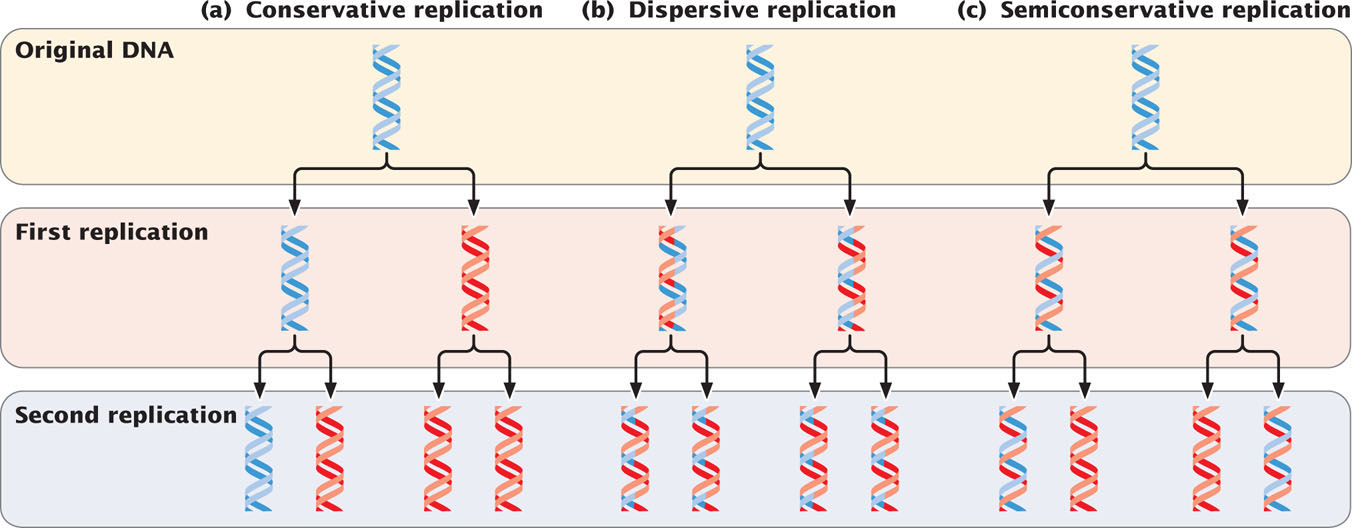
Initially, three models were proposed for DNA replication. In conservative replication (Figure 12.1a), the entire double-stranded DNA molecule serves as a template for a whole new molecule of DNA, and the original DNA molecule is fully conserved during replication. In dispersive replication (Figure 12.1b), both nucleotide strands break down (disperse) into fragments, which serve as templates for the synthesis of new DNA fragments, and then somehow reassemble into two complete DNA molecules. In this model, each resulting DNA molecule is interspersed with fragments of old and new DNA; none of the original molecule is conserved. Semiconservative replication (Figure 12.1c) is intermediate between these two models; the two nucleotide strands unwind and each serves as a template for a new DNA molecule.
327
These three models allow different predictions to be made about the distribution of original DNA and newly synthesized DNA after replication. With conservative replication, after one round of replication, 50% of the molecules would consist entirely of the original DNA and 50% would consist entirely of new DNA. After a second round of replication, 25% of the molecules would consist entirely of the original DNA and 75% would consist entirely of new DNA. With each additional round of replication, the proportion of molecules with new DNA would increase, although the number of molecules with the original DNA would remain constant. Dispersive replication would always produce hybrid molecules, containing some original and some new DNA, but the proportion of new DNA within the molecules would increase with each replication event. In contrast, with semiconservative replication, one round of replication would produce two hybrid molecules, each consisting of half original DNA and half new DNA. After a second round of replication, half the molecules would be hybrid, and the other half would consist of new DNA only. Additional rounds of replication would produce more and more molecules consisting entirely of new DNA, and a few hybrid molecules would persist.
Meselson and Stahl’s Experiment
To determine which of the three models of replication applied to E. coli cells, Matthew Meselson and Franklin Stahl needed a way to distinguish old and new DNA. They did so by using two isotopes of nitrogen, 14N (the common form) and 15N (a rare, heavy form). Meselson and Stahl grew a culture of E. coli in a medium that contained 15N as the sole nitrogen source; after many generations, all the E. coli cells had 15N incorporated into all of the purine and pyrimidine bases of their DNA (see Figure 10.10). Meselson and Stahl took a sample of these bacteria, switched the rest of the bacteria to a medium that contained only 14N, and then took additional samples of bacteria over the next few cellular generations. In each sample, the bacterial DNA that was synthesized before the change in medium contained 15N and was relatively heavy, whereas any DNA synthesized after the switch contained 14N and was relatively light.
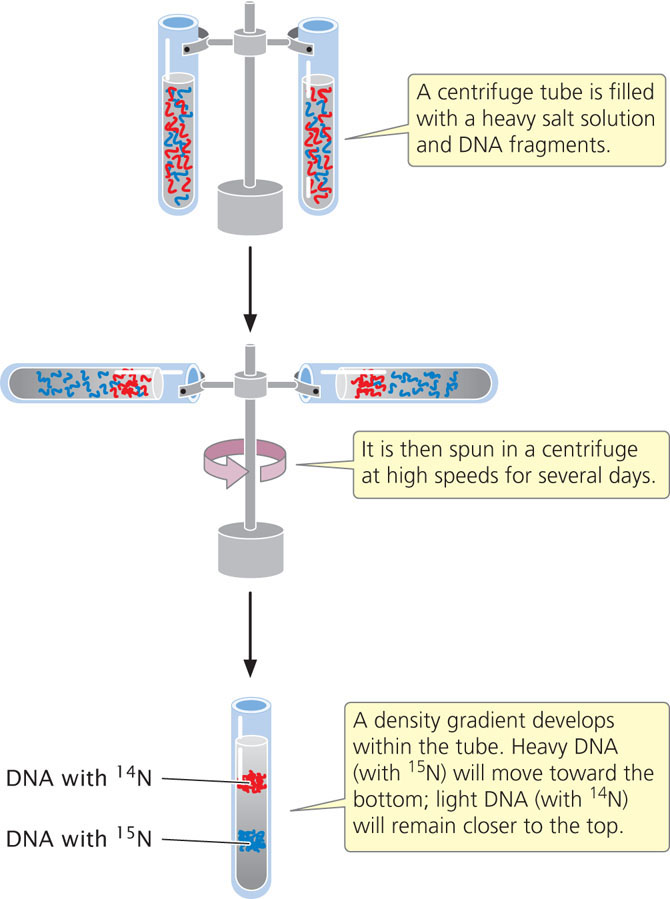
Meselson and Stahl distinguished between the heavy 15N-laden DNA and the light 14N-containing DNA with the use of equilibrium density gradient centrifugation (Figure 12.2).
328
In this technique, a centrifuge tube is filled with a heavy salt solution and a substance of unknown density—in this case, DNA fragments. The tube is then spun in a centrifuge at high speeds. After several days of spinning, a gradient of density develops within the tube, with high density at the bottom and low density at the top. The density of the DNA fragments matches that of the salt: light molecules rise and heavy molecules sink.
Meselson and Stahl found that DNA from bacteria grown only on medium containing 15N produced a single band at the position expected of DNA containing only 15N (Figure 12.3a). DNA from bacteria transferred to the medium with 14N and allowed one round of replication also produced a single band but at a position intermediate between that expected of DNA containing only 15N and that expected of DNA containing only 14N (Figure 12.3b). This result is inconsistent with the conservative replication model, which predicts one heavy band (the original DNA molecules) and one light band (the new DNA molecules). A single band of intermediate density is predicted by both the semiconservative and the dispersive models.
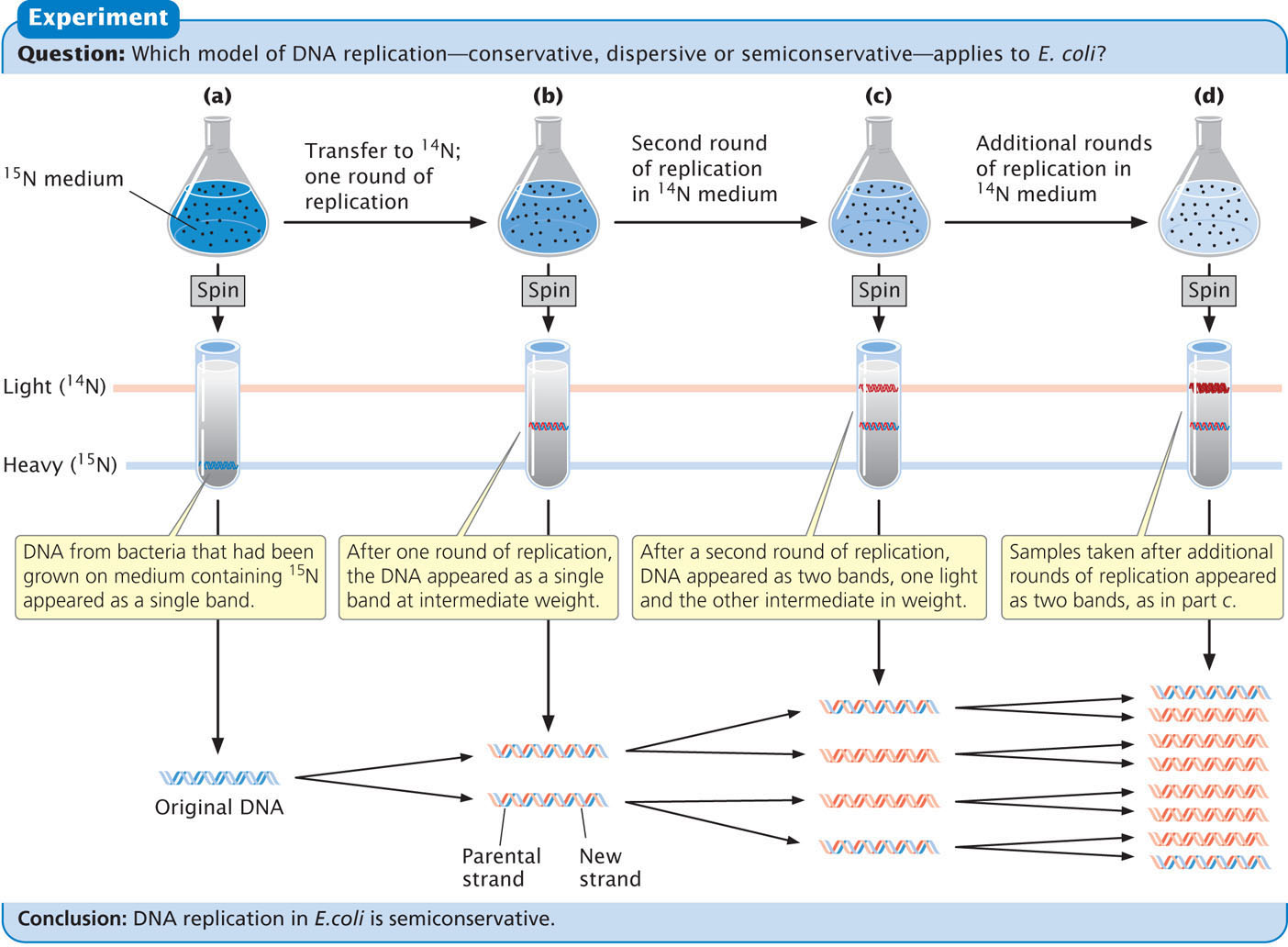
To distinguish between these two models, Meselson and Stahl grew the bacteria in medium containing 14N for a second generation. After a second round of replication in medium with 14N, two bands of equal intensity appeared, one in the intermediate position and the other at the position expected of DNA containing only 14N (Figure 12.3c). All samples taken after additional rounds of replication produced the same two bands, and the band representing light DNA became progressively stronger (Figure 12.3d). Meselson and Stahl’s results were exactly as expected for semiconservative replication and are incompatible with those predicated for both conservative and dispersive replication.  TRY PROBLEM 22
TRY PROBLEM 22
329
CONCEPTS
Replication is semiconservative: each DNA strand serves as a template for the synthesis of a new DNA molecule. Meselson and Stahl convincingly demonstrated that replication in E. coli is semiconservative.
 CONCEPT CHECK 1
CONCEPT CHECK 1
How many bands of DNA would be expected in Meselson and Stahl’s experiment after two rounds of conservative replication?
Modes of Replication
After Meselson and Stahl’s work, investigators confirmed that other organisms also use semiconservative replication. No evidence was found for conservative or dispersive replication. There are, however, several different ways in which semiconservative replication can take place, differing principally in the nature of the template DNA—that is, whether it is linear or circular.
Individual units of replication are called replicons, each of which contains a replication origin. Replication starts at the origin and continues until the entire replicon has been replicated. Bacterial chromosomes have a single replication origin, whereas eukaryotic chromosomes contain many.
Theta Replication
A common type of replication that takes place in circular DNA, such as that found in E. coli and other bacteria, is called theta replication (Figure 12.4a) because it generates a structure that resembles the Greek letter theta (θ). In this and all subsequent figures in this chapter, the original (template) strand of DNA is shown in gray and the newly synthesized strand of DNA is shown in red.
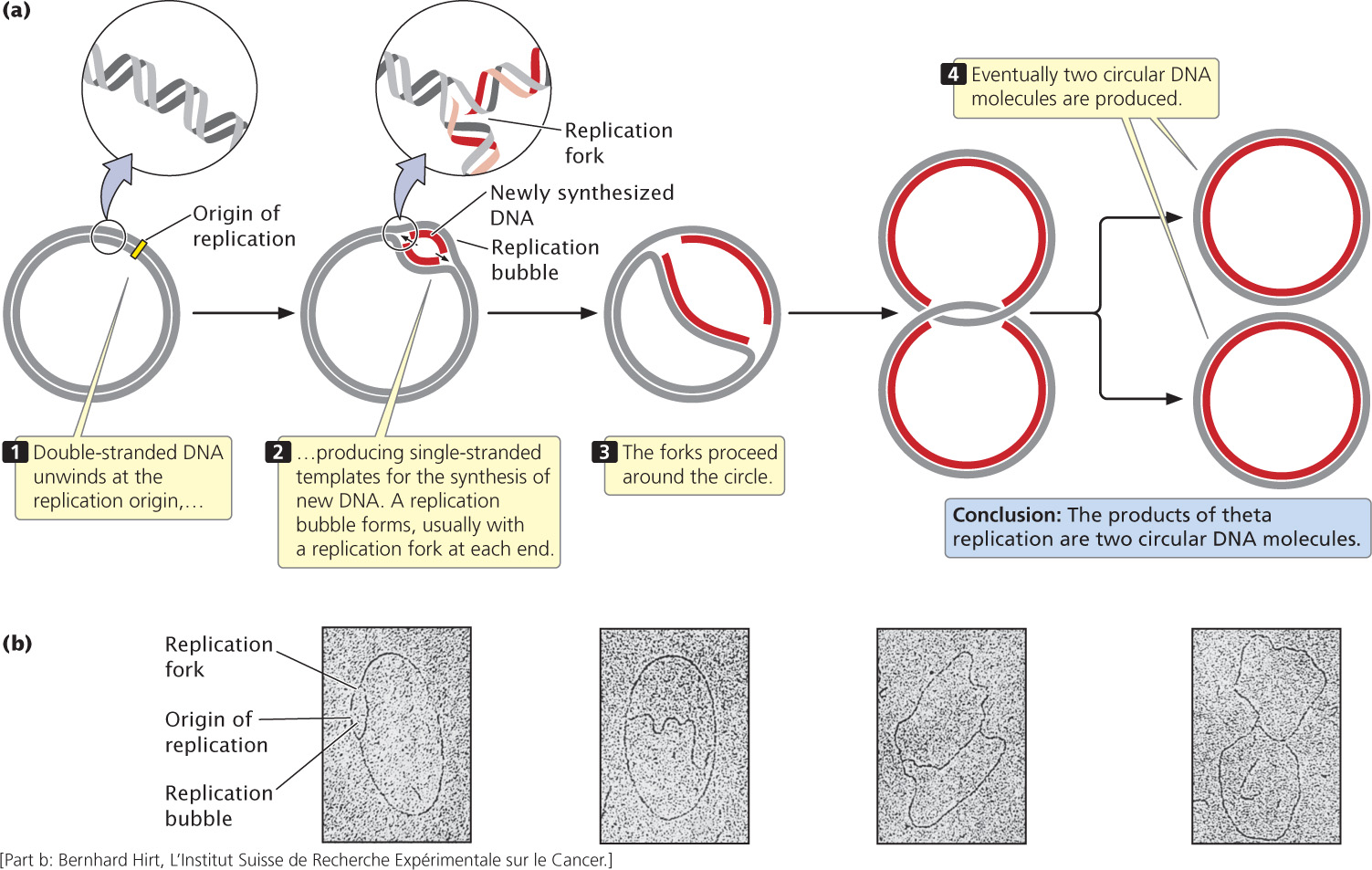
In theta replication, double-stranded DNA begins to unwind at the replication origin, producing single-stranded nucleotide strands that then serve as templates on which new DNA can be synthesized. The unwinding of the double helix generates a loop, termed a replication bubble. Unwinding may be at one or both ends of the bubble, making it progressively larger. DNA replication on both of the template strands is simultaneous with unwinding. The point of unwinding, where the two single nucleotide strands separate from the double-stranded DNA helix, is called a replication fork.
If there are two replication forks, one at each end of the replication bubble, the forks proceed outward in both directions in a process called bidirectional replication, simultaneously unwinding and replicating the DNA until they eventually meet. If a single replication fork is present, it proceeds around the entire circle. Both bidirectional and unidirectional replication produce two complete circular DNA molecules, each consisting of one old and one new nucleotide strand.
330
John Cairns provided the first visible evidence of theta replication in 1963 by growing bacteria in the presence of radioactive nucleotides. After replication, each DNA molecule consisted of one “hot” (radioactive) strand and one “cold” (nonradioactive) strand. Cairns isolated DNA from the bacteria after replication, placed it on an electron-microscope grid, and then covered it with a photographic emulsion. Radioactivity present in the sample exposed the emulsion and produced a picture of the molecule (called an autoradiograph), similar to the way in which light exposes a photographic film. Because the newly synthesized DNA contained radioactive nucleotides, Cairns was able to produce an electron micrograph of the replication process, similar to those shown in Figure 12.4b.
Rolling-Circle Replication
Another form of replication, called rolling-circle replication (Figure 12.5), takes place in some viruses and in the F factor (a small circle of extrachromosomal DNA that controls mating, discussed in Chapter 9) of E. coli. This form of replication is initiated by a break in one of the nucleotide strands that creates a 3′-OH group and a 5′-phosphate group. New nucleotides are added to the 3′ end of the broken strand, with the inner (unbroken) strand used as a template. As new nucleotides are added to the 3′ end, the 5′end of the broken strand is displaced from the template, rolling out like thread being pulled off a spool. The 3′ end grows around the circle, giving rise to the name rolling-circle.
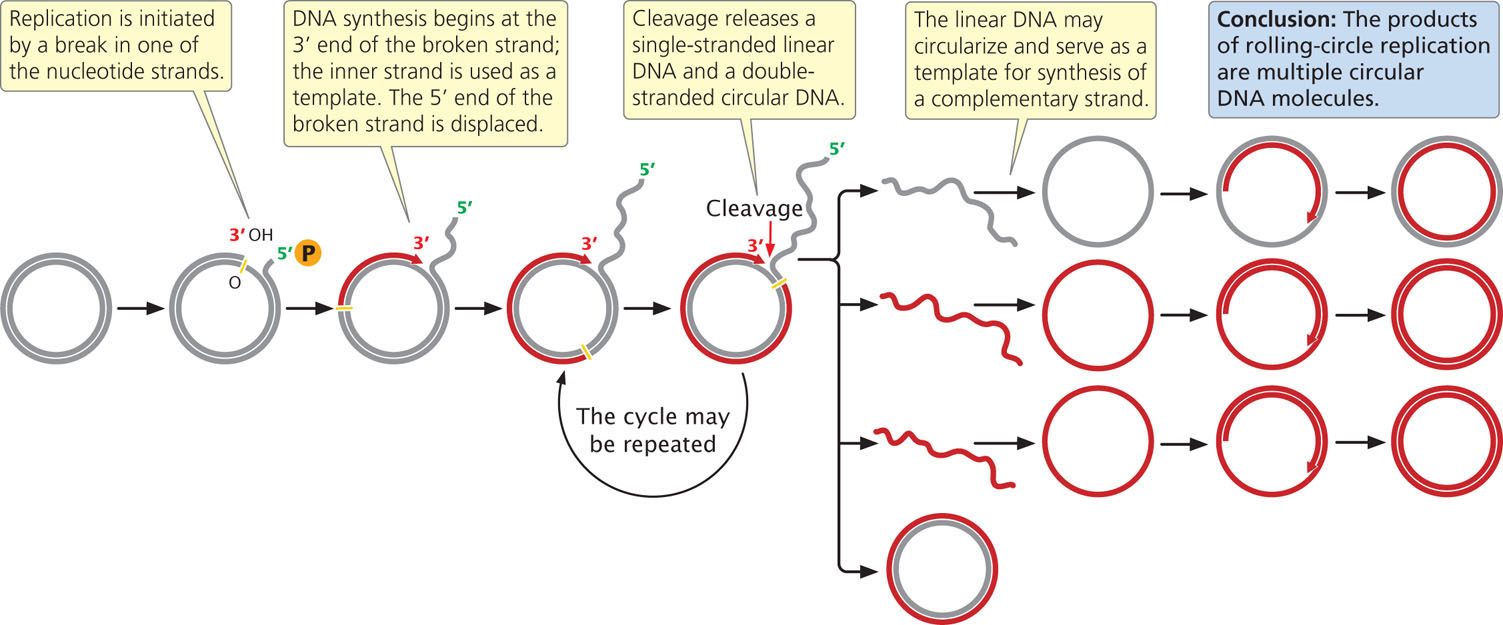
The replication fork may continue around the circle a number of times, producing several linked copies of the same sequence. With each revolution around the circle, the growing 3′ end displaces the nucleotide strand synthesized in the preceding revolution. Eventually, the linear DNA molecule is cleaved from the circle, resulting in a double-stranded circular DNA molecule and a single-stranded linear DNA molecule. The linear molecule circularizes either before or after serving as a template for the synthesis of a complementary strand.
Linear Eukaryotic Replication
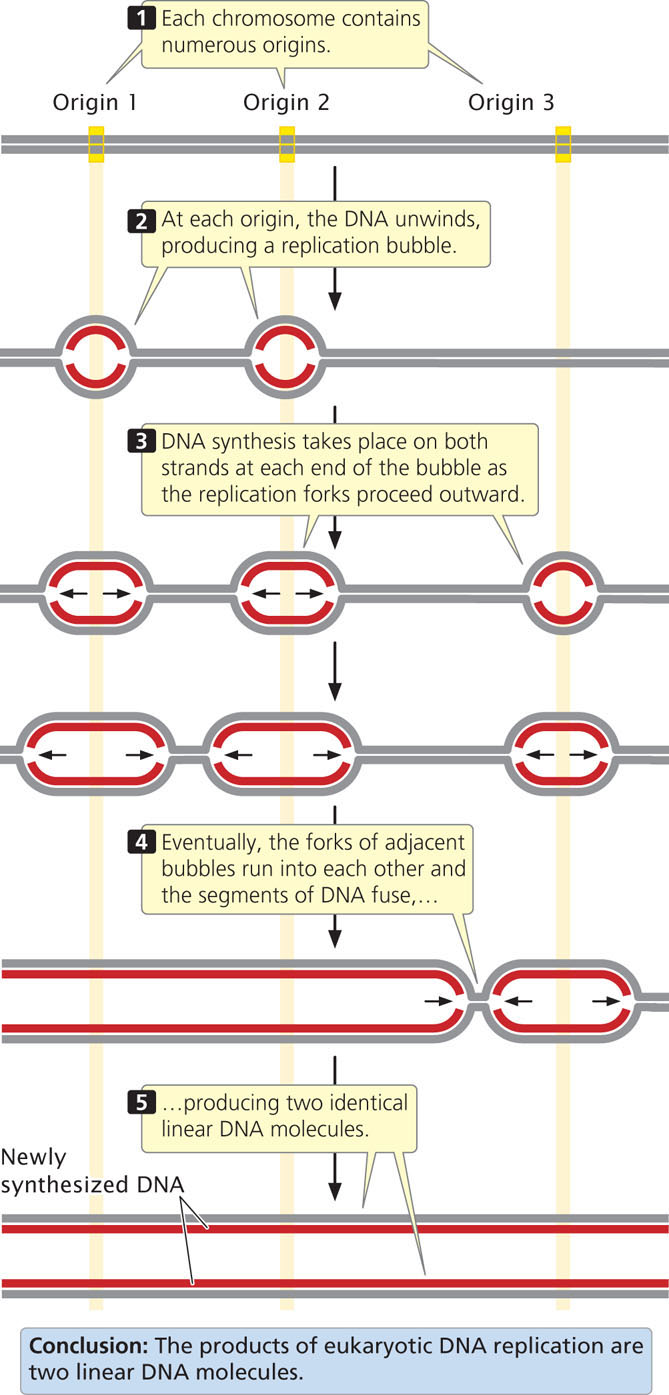
Circular DNA molecules that undergo theta or rolling-circle replication have a single origin of replication. Because of the limited size of these DNA molecules, replication starting from one origin can traverse the entire chromosome in a reasonable amount of time. The large linear chromosomes in eukaryotic cells, however, contain far too much DNA to be replicated speedily from a single origin. Eukaryotic replication proceeds at a rate ranging from 500 to 5000 nucleotides per minute at each replication fork (considerably slower than bacterial replication). Even at 5000 nucleotides per minute at each fork, DNA synthesis starting from a single origin would require 7 days to replicate a typical human chromosome consisting of 100 million base pairs of DNA. The replication of eukaryotic chromosomes actually takes place in a matter of minutes or hours, not days. This rate is possible because replication initiates at thousands of origins.
Typical eukaryotic replicons are from 20,000 to 300,000 base pairs in length (Table 12.1). At each replication origin, the DNA unwinds and produces a replication bubble. Replication takes place on both strands at each end of the bubble, with the two replication forks spreading outward. Eventually, the replication forks of adjacent replicons run into each other, and the replicons fuse to form long stretches of newly synthesized DNA (Figure 12.6). Replication and fusion of all the replicons leads to two identical DNA molecules. Important features of theta replication, rolling-circle replication, and linear eukaryotic replication are summarized in Table 12.2.  TRY PROBLEM 23
TRY PROBLEM 23
| Organism | Number of Replication Origins | Average Length of Replicon (bp) |
|---|---|---|
| Escherichia coli (bacterium) | 1 | 4,600,000 |
| Saccharomyces cerevisiae (yeast) | 500 | 40,000 |
| Drosophila melanogaster (fruit fly) | 3,500 | 40,000 |
| Xenopus laevis (frog) | 15,000 | 200,000 |
| Mus musculus (mouse) | 25,000 | 150,000 |
| Source: Data from B. L. Lewin, Genes V (Oxford: Oxford University Press, 1994), p. 536. | ||
| Replication Model | DNA Template | Breakage of Nucleotide Strand | Number of Replicons | Unidirectional or Bidirectional | Products |
|---|---|---|---|---|---|
| Theta | Circular | No | 1 | Unidirectional or bidirectional | Two circular molecules |
| Rolling circle | Circular | Yes | 1 | Unidirectional | One circular molecule and one linear molecule that may circularize |
| Linear eukaryotic | Linear | No | Many | Bidirectional | Two linear molecules |
331
CONCEPTS
Theta replication, rolling-circle replication, and linear replication differ with respect to the initiation and progression of replication, but all produce new DNA molecules by semiconservative replication.
 CONCEPT CHECK 2
CONCEPT CHECK 2
Which type of replication requires a break in the nucleotide strand to get started?
- Theta replication
- Rolling-circle replication
- Linear eukaryotic replication
- All of the above
332
Requirements of Replication
Although the process of replication includes many components, they can be combined into three major groups:
- 1. A template consisting of single-stranded DNA
- 2. Raw materials (substrates) to be assembled into a new nucleotide strand
- 3. Enzymes and other proteins that “read” the template and assemble the substrates into a DNA molecule
Because of the semiconservative nature of DNA replication, a double-stranded DNA molecule must unwind to expose the bases that act as a template for the assembly of new polynucleotide strands, which will be complementary and antiparallel to the template strands. The raw materials from which new DNA molecules are synthesized are deoxyribonucleoside triphosphates (dNTPs), each consisting of a deoxyribose sugar and a base (a nucleoside) attached to three phosphate groups (Figure 12.7a). In DNA synthesis, nucleotides are added to the 3′-OH group of the growing nucleotide strand (Figure 12.7b). The 3′-OH group of the last nucleotide on the strand attacks the 5′-phosphate group of the incoming dNTP. Two phosphate groups are cleaved from the incoming dNTP, and a phosphodiester bond is created between the two nucleotides.
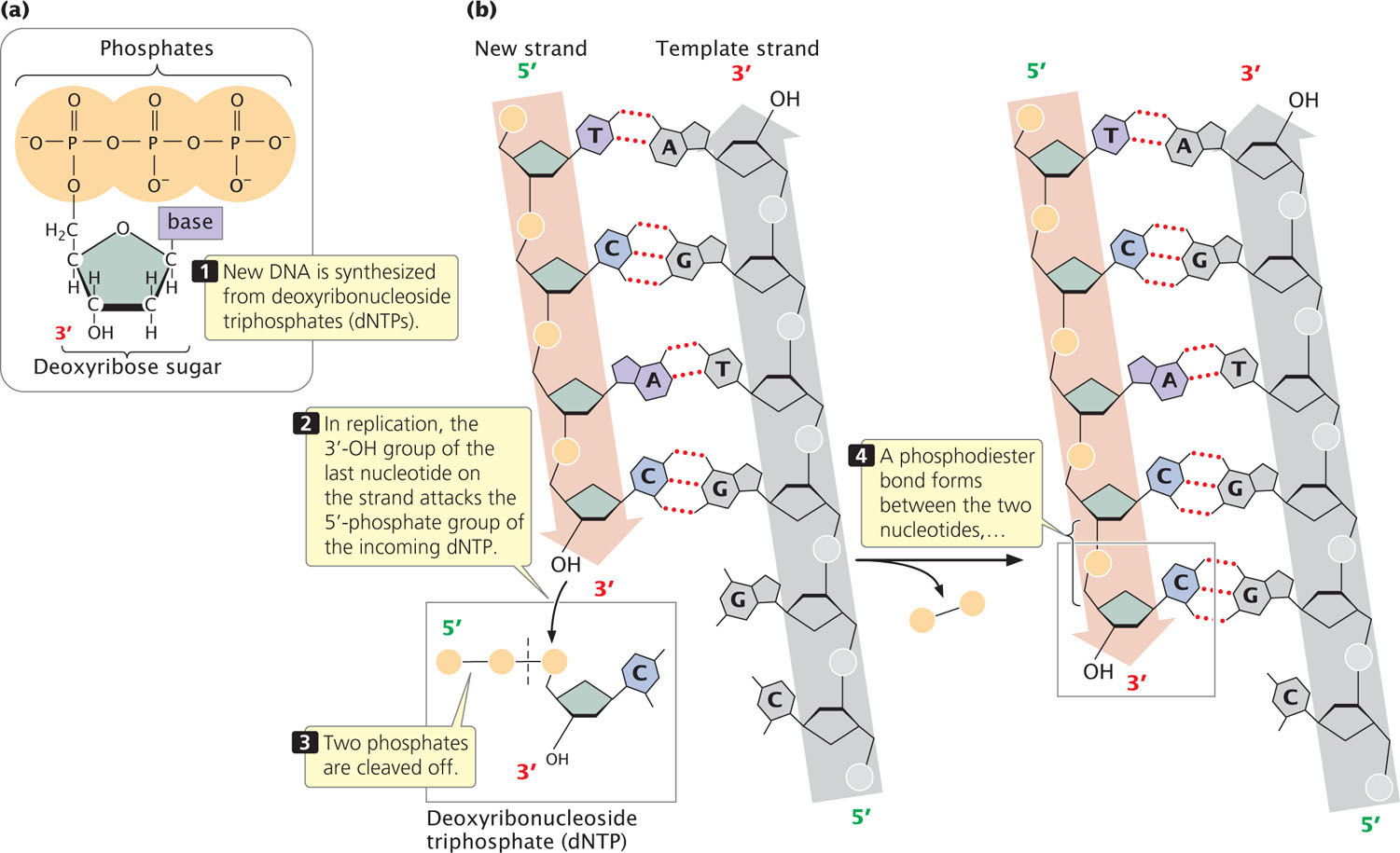
The newly synthesized strand is complementary and antiparallel to the template strand; the two strands are held together by hydrogen bonds (represented by red dotted lines) between the bases.
DNA synthesis does not happen spontaneously. Rather, it requires a host of enzymes and proteins that function in a coordinated manner. We will examine this complex array of proteins and enzymes as we consider the replication process in more detail.
CONCEPTS
DNA synthesis requires a single-stranded DNA template, deoxyribonucleoside triphosphates, a growing nucleotide strand, and a group of enzymes and proteins.
Direction of Replication
In DNA synthesis, new nucleotides are joined one at a time to the 3′ end of the newly synthesized strand. DNA polymerases, the enzymes that synthesize DNA, can add nucleotides only to the 3′ end of the growing strand (not the 5′ end), and so new DNA strands always elongate in the same 5′-to-3′ direction (5′→3′). Because the two single-stranded DNA templates are antiparallel and strand elongation is always 5′→3′, if synthesis on one template proceeds from, say, right to left, then synthesis on the other template must proceed in the opposite direction, from left to right (Figure 12.8). As DNA unwinds during replication, the antiparallel nature of the two DNA strands means that one template is exposed in the 5′→3′ direction and the other template is exposed in the 3′→5′ direction. So how can synthesis take place simultaneously on both strands at the fork?
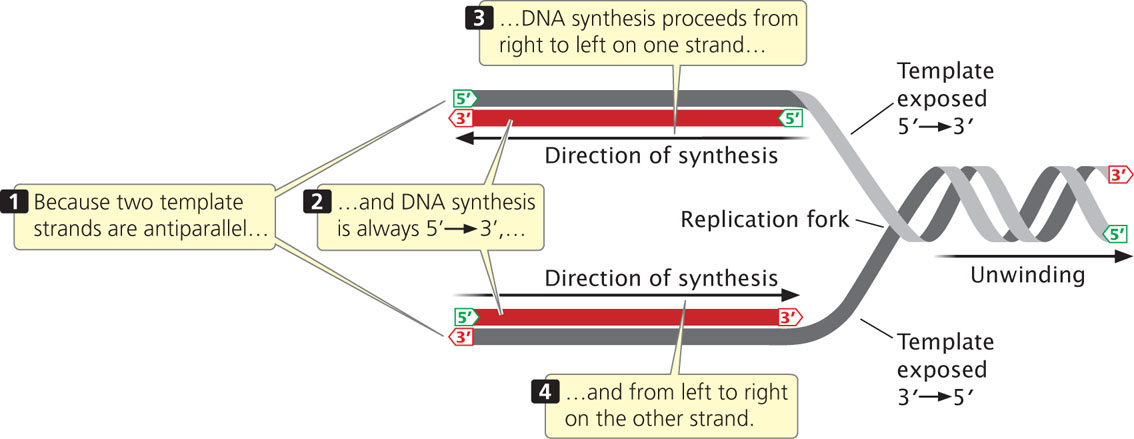
DNA replication at a single replication fork begins when a double-stranded DNA molecule unwinds to provide two single-strand templates.
Continuous and Discontinuous Replication
As the DNA unwinds, the template strand that is exposed in the 3′→5′ direction (the lower strand in Figures 12.8 and 12.9) allows the new strand to be synthesized continuously, in the 5′→3′ direction. This new strand, which undergoes continuous replication, is called the leading strand.
333
The other template strand is exposed in the 5′→3′ direction (the upper strand in Figures 12.8 and 12.9). After a short length of the DNA has been unwound, synthesis must proceed 5′→3′; that is, in the direction opposite that of unwinding (Figure 12.9). Because only a short length of DNA needs to be unwound before synthesis on this strand gets started, the replication machinery soon runs out of template. By that time, more DNA has unwound, providing new template at the 5′ end of the new strand. DNA synthesis must start anew at the replication fork and proceed in the direction opposite that of the movement of the fork until it runs into the previously replicated segment of DNA. This process is repeated again and again, so synthesis of this strand is in short, discontinuous bursts. The newly made strand that undergoes discontinuous replication is called the lagging strand.
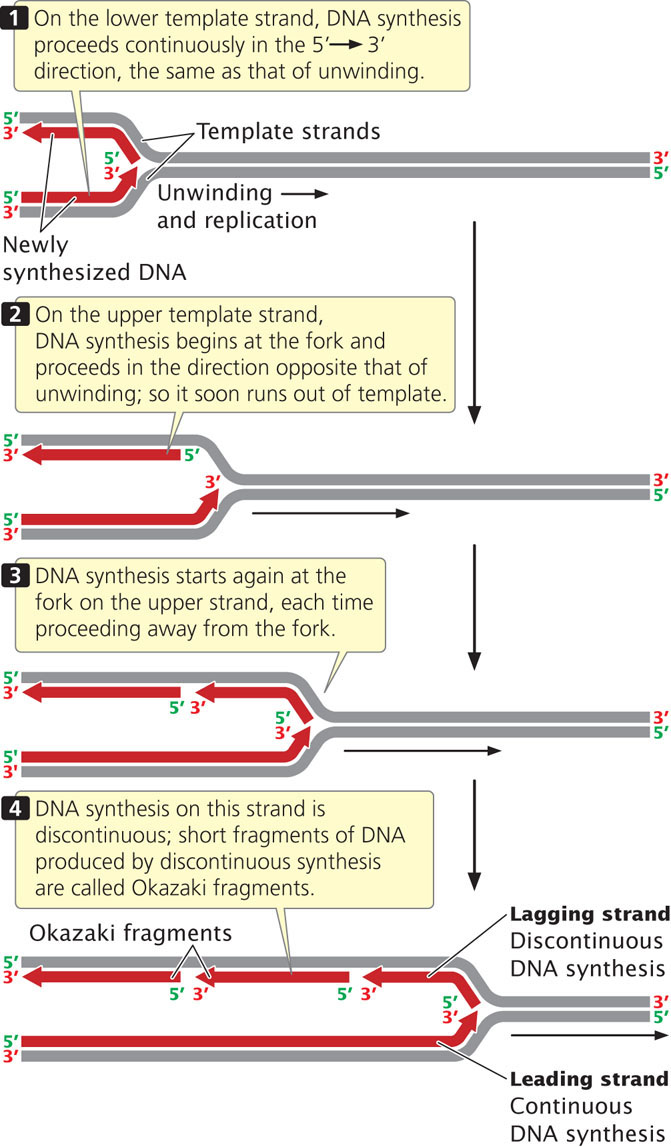
Okazaki Fragments
The short lengths of DNA produced by discontinuous replication of the lagging strand are called Okazaki fragments, after Reiji Okazaki, who discovered them. In bacterial cells, each Okazaki fragment ranges from about 1000 to 2000 nucleotides in length; in eukaryotic cells, they are about 100 to 200 nucleotides long. Okazaki fragments on the lagging strand are linked together to create a continuous new DNA molecule. To see how replication occurs continuously on one strand and discontinuously on the other, view  Animation 12.1.
Animation 12.1.
CONCEPTS
All DNA synthesis is 5′→3′, meaning that new nucleotides are always added to the 3′ end of the growing nucleotide strand. At each replication fork, synthesis of the leading strand proceeds continuously and that of the lagging strand proceeds discontinuously.
 CONCEPT CHECK 3
CONCEPT CHECK 3
Discontinuous replication is a result of which property of DNA?
- Complementary bases
- Charged phosphate group
- Antiparallel nucleotide strands
- Five-carbon sugar
334
CONNECTING CONCEPTS: The Direction of Synthesis in Different Models of Replication
Let’s relate the direction of DNA synthesis to the modes of replication examined earlier. In the theta model (Figure 12.10a), the DNA unwinds at one particular location, the origin, and a replication bubble is formed. If the bubble has two forks, one at each end, synthesis takes place simultaneously at both forks (bidirectional replication). At each fork, synthesis on one of the template strands proceeds in the same direction as that of unwinding; this newly replicated strand is the leading strand with continuous replication. On the other template strand, synthesis proceeds in the direction opposite that of unwinding; this newly synthesized strand is the lagging strand with discontinuous replication. Focus on just one of the template strands within the bubble. Notice that synthesis on this template strand is continuous at one fork but discontinuous at the other. This difference arises because DNA synthesis is always in the same direction (5′→3′), but the two forks are moving in opposite directions.
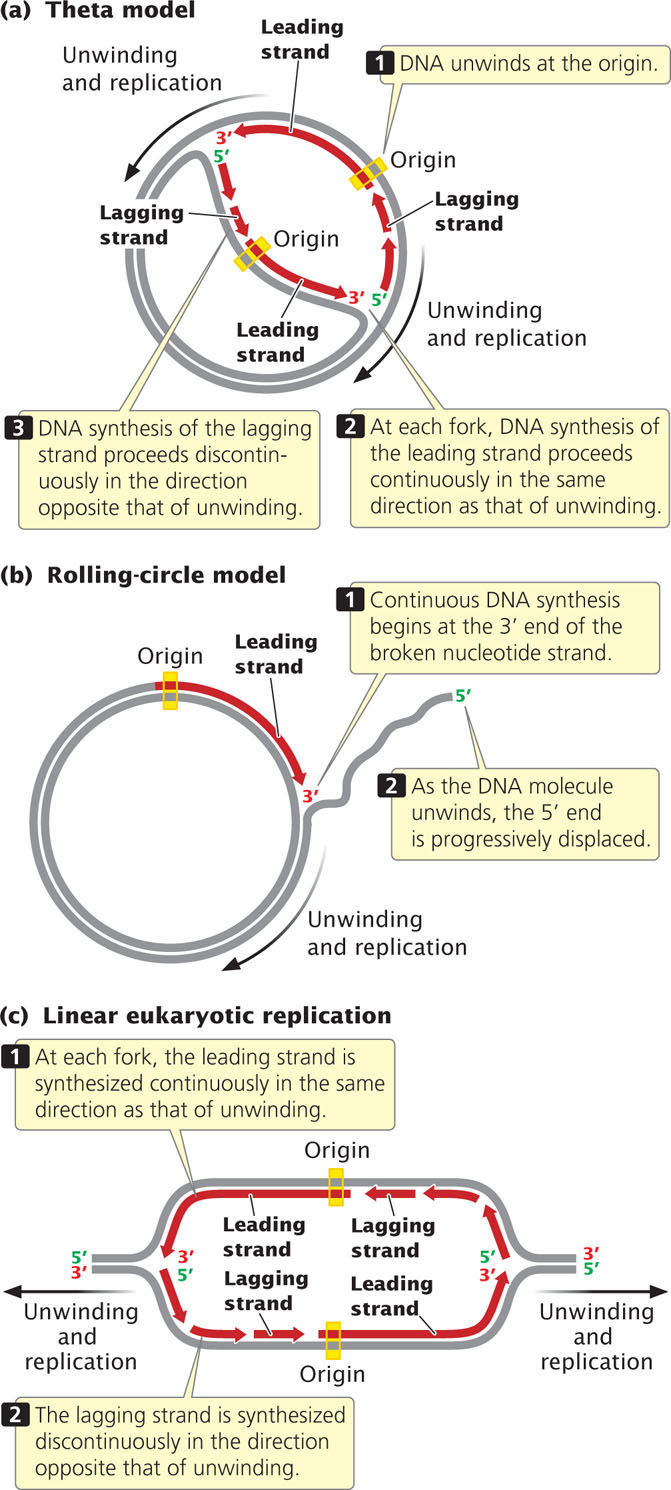
Replication in the rolling-circle model (Figure 12.10b) is somewhat different, because there is no replication bubble. Replication begins at the 3′ end of the broken nucleotide strand. Continuous replication takes place on the circular template as new nucleotides are added to this 3′ end.
The replication of linear molecules of DNA, such as those found in eukaryotic cells, produces a series of replication bubbles (Figure 12.10c). DNA synthesis in these bubbles is the same as that in the single replication bubble of the theta model; it begins at the center of each replication bubble and proceeds at two forks, one at each end of the bubble. At both forks, synthesis of the leading strand proceeds in the same direction as that of unwinding, whereas synthesis of the lagging strand proceeds in the direction opposite that of unwinding.  TRY PROBLEM 25a-c
TRY PROBLEM 25a-c