Challenge Questions
Section 13.3
Question 13.37
Many genes in both bacteria and eukaryotes contain numerous sequences that potentially cause pauses or premature terminations of transcription. Nevertheless, the transcription of these genes within a cell normally produces multiple RNA molecules thousands of nucleotides long without pausing or terminating prematurely. However, when a single round of transcription takes place on such templates in a test tube, RNA synthesis is frequently interrupted by pauses and premature terminations, which reduce the rate at which transcription takes place and frequently shortens the length of the mRNA molecules produced. Most pauses and premature terminations occur when RNA polymerase temporarily backtracks (i.e., backs up) for one or two nucleotides along the DNA. Experimental findings have demonstrated that most transcriptional delays and premature terminations disappear if several RNA polymerases are simultaneously transcribing the DNA molecule. Propose an explanation for faster transcription and longer mRNA when the template DNA is being transcribed by multiple RNA polymerases.
Section 13.4
Question 13.38
 Enhancers are sequences that affect the initiation of the transcription of genes that are hundreds or thousands of nucleotides away. Transcriptional activator proteins that bind to enhancers usually interact directly with transcription factors at promoters by causing the intervening DNA to loop out. An enhancer of bacteriophage T4 does not function by looping of the DNA (D. R. Herendeen et al. 1992. Science 256: 1298-1303). Propose some additional mechanisms (other than DNA looping) by which this enhancer might affect transcription at a gene thousands of nucleotides away.
Enhancers are sequences that affect the initiation of the transcription of genes that are hundreds or thousands of nucleotides away. Transcriptional activator proteins that bind to enhancers usually interact directly with transcription factors at promoters by causing the intervening DNA to loop out. An enhancer of bacteriophage T4 does not function by looping of the DNA (D. R. Herendeen et al. 1992. Science 256: 1298-1303). Propose some additional mechanisms (other than DNA looping) by which this enhancer might affect transcription at a gene thousands of nucleotides away.
Question 13.39
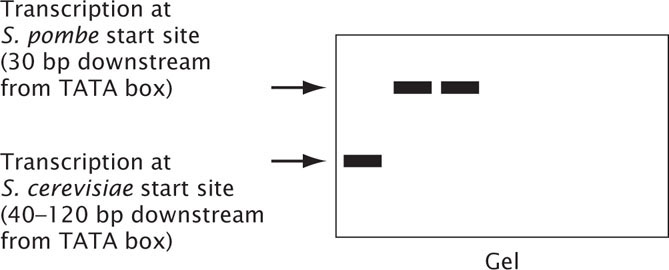
 The locations of the TATA box in two species of yeast, Saccharomyces pombe and Saccharomyces cerevisiae, differ dramatically. The TATA box of S. pombe is about 30 nucleotides upstream of the start site, similar to the location in most other eukaryotic cells. However, the TATA box of S. cerevisiae is 40 to 120 nucleotides upstream of the start site.
The locations of the TATA box in two species of yeast, Saccharomyces pombe and Saccharomyces cerevisiae, differ dramatically. The TATA box of S. pombe is about 30 nucleotides upstream of the start site, similar to the location in most other eukaryotic cells. However, the TATA box of S. cerevisiae is 40 to 120 nucleotides upstream of the start site.
To better understand what sets the start site in these organisms, researchers at Stanford University conducted a series of experiments to determine which components of the transcription apparatus of these two species could be interchanged (Y. Li et al. 1994. Science 263:805–807). In these experiments, different transcription factors and RNA polymerases were switched in S. pombe and S. cerevisiae, and the effects of the switch on the level of RNA synthesis and on the start point of transcription were observed. The results from one set of experiments are shown in the table below. Components cTFIIB, cTFIIE, cTFIIF, cTFIIH are transcription factors from S. cerevisisae. Components pTFIIB, pTFIIE, pTFIIF, pTFIIH are transcription factors from S. pombe. Components cPol II and pPol II are RNA polymerase II from S. cerevisiae and S. pombe, respectively. The table indicates whether the component was present (+) or missing (−) in the experiment. In the accompanying gel, the presence of a band indicates that RNA was produced and the position of the band indicates whether it was the length predicated when transcription begins 30 bp downstream from the TATA box or from 40 to 120 bp downstream from the TATA box.
| Experiment | |||||||
|---|---|---|---|---|---|---|---|
| Components | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| cTFIIE | + | − | + | + | + | + | − |
| cTFIIH + cTFIIF | + | − | + | + | + | − | + |
| cTFIIB + cPol II | + | − | − | − | − | − | − |
| pPol II | − | + | + | + | − | + | + |
| pTFIIB | − | + | + | − | + | + | + |
| pTFIIE + pTFIIH + pTFIIF | − | + | − | − | − | − | − |
381
- a. What conclusion can you draw from these data about what components determine the start site for transcription?
- b. What conclusions can you draw about the interactions of the different components of the transcription apparatus?
 S. pombe.[Steve Gschmeissner/Photo Researchers.]
S. pombe.[Steve Gschmeissner/Photo Researchers.] - c. Propose a mechanism for why the start site for transcription in S. pombe is about 30 bp downstream from the TATA box, whereas the start site for transcription in S. cerevisiae is 40 to 120 bp downstream from the TATA box.
Question 13.40
 Glenn Croston and his colleagues studied the relation between chromatin structure and transcription activity. In one set of experiments, they measured the level of in vitro transcription of a Drosophila gene by RNA polymerase II with the use of DNA and various combinations of histone proteins (G. E. Croston et al. 1991. Science 251:643-649).
Glenn Croston and his colleagues studied the relation between chromatin structure and transcription activity. In one set of experiments, they measured the level of in vitro transcription of a Drosophila gene by RNA polymerase II with the use of DNA and various combinations of histone proteins (G. E. Croston et al. 1991. Science 251:643-649).
First, they measured the level of transcription for naked DNA, with no associated histone proteins. Then, they measured the level of transcription after nucleosome octamers (without H1) were added to the DNA. The addition of the octamers caused the level of transcription to drop by 50%. When both the nucleosome octamers and the H1 proteins were added to the DNA, transcription was greatly repressed, dropping to less than 1% of that obtained with naked DNA, as shown in the table below.
GAL4-VP16 is a protein that binds to the DNA of certain eukaryotic genes. When GAL4-VP16 is added to DNA, the level of RNA polymerase II transcription is greatly elevated.
| Treatment | Relative amount of transcription |
|---|---|
| Naked DNA | 100 |
| DNA + octamers | 50 |
| DNA + octamers + H1 | <1 |
| DNA + GAL4-VP16 | 1000 |
| DNA + octamers + GAL4-VP16 | 1000 |
| DNA + octamers + H1 + GAL4-VP16 | 1000 |
Even in the presence of the H1 protein, GAL4-VP16 stimulates high levels of transcription.
Propose a mechanism for how the H1 protein represses transcription and how GAL4-VP16 overcomes this repression. Explain how your proposed mechanism would produce the results obtained in these experiments.
Go to your  to find additional learning resources and the Suggested Readings for this chapter.
to find additional learning resources and the Suggested Readings for this chapter.
382