Application Questions and Problems
Section 15.1
Question 15.16
Sydney Brenner isolated Salmonella typhimurium mutants that were implicated in the biosynthesis of tryptophan and would not grow on minimal medium. When these mutants were tested on minimal medium to which one of four compounds (indole glycerol phosphate, indole, anthranilic acid, and tryptophan) had been added, the growth responses shown in the following table were obtained.
| Mutant | Minimal medium | Anthranilic acid | Indole glycerol phosphate | Indole | Tryptophan |
|---|---|---|---|---|---|
| trp-1 | − | − | − | − | + |
| trp-2 | − | − | + | + | + |
| trp-3 | − | − | − | + | + |
| trp-4 | − | − | + | + | + |
| trp-6 | − | − | − | − | + |
| trp-7 | − | − | − | − | + |
| trp-8 | − | + | + | + | + |
| trp-9 | − | − | − | − | + |
| trp-10 | − | − | − | − | + |
| trp-11 | − | − | − | − | + |
Give the order of indole glycerol phosphate, indole, anthranilic acid, and tryptophan in a biochemical pathway leading to the synthesis of tryptophan. Indicate which step in the pathway is affected by each of the mutations.
Question 15.17
Compounds I, II, and III are in the following biochemical pathway:
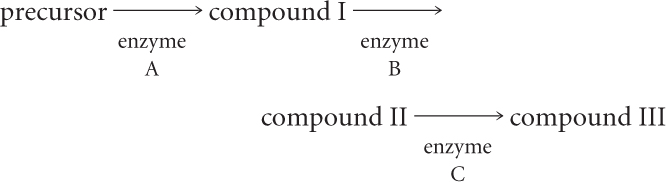
Mutation a inactivates enzyme A, mutation b inactivates enzyme B, and mutation c inactivates enzyme C. Mutants, each having one of these defects, were tested on minimal medium to which compound I, II, or III was added. Fill in the results expected of these tests by placing a plus sign (+) for growth or a minus sign (−) for no growth in the table below.
| Minimal medium to which is added | |||
|---|---|---|---|
| Strain with mutation | Compound | Compound | Compound |
| I | II | III | |
| a | |||
| b | |||
| c | |||
Section 15.2
Question 15.18
A geneticist conducts the experiment outlined in Figure 15.8, but this time she combines guanine nucleotides (instead of uracil) with polynucleotide phosphorylase. Radioactively labeled protein should appear in which tube?
Question 15.19
For the experiment outlined in Figure 15.8, could Nirenberg and Mattaei have substituted RNA polymerase instead of polynucleotide phosphorylase without otherwise modifying the experiment? Why or why not?
439
Question 15.20
Assume that the number of different types of bases in RNA is four. What would be the minimum codon size (number of nucleotides) required to specify all amino acids if the number of different types of amino acids in proteins were: (a) 2, (b) 8, (c) 17, (d) 45, (e) 75?
Question 15.21
How many codons would be possible in a triplet code if only three bases (A, C, and U) were used?
Question 15.22
Referring to the genetic code presented in Figure 15.10, give the amino acids specified by the following bacterial mRNA sequences.
- a. 5′–AUGUUUAAAUUUAAAUUUUGA–3′
- b. 5′–AGGGAAAUCAGAUGUAUAUAUAUAUAUGA–3′
- c. 5′–UUUGGAUUGAGUGAAACGAUG GAUGAAAGAUUUCUCGCUUGA–3′
- d. 5′–GUACUAAGGAGGUUGUAUGGG UUAGGGGACAUCAUUUUGA–3′
Question 15.23
A nontemplate strand on bacterial DNA has the following base sequence. What amino acid sequence will be encoded by this sequence?
5′–ATGATACTAAGGCCC–3′
Question 15.24
The following amino acid sequence is found in a tripeptide: Met-Trp-His. Give all possible nucleotide sequences on the mRNA, on the template strand of DNA, and on the nontemplate strand of DNA that can encode this tripeptide.
Question 15.25
How many different mRNA sequences can encode a polypeptide chain with the amino acid sequence MetLeu-Arg? (Be sure to include the stop codon.)
Question 15.26
A series of tRNAs have the following anticodons. Consider the wobble rules listed in Table 15.2 and give all possible codons with which each tRNA can pair.
- a. 5′–GGC–3′
- b. 5′–AAG–3′
- c. 5′–IAA–3′
- d. 5′–UGG–3′
- e. 5′–CAG–3′
Question 15.27
A researcher creates random copolymers of three nucleotides by mixing polynucletide phosphorylase with guanine and adenine nucleotides in a ratio of 5 guanine nucleotides to 1 adenine. Give the different copolymers produced and their theoretical proportions.
Question 15.28
Assume that the nucleotide at the 5′ end of the first tRNA’s anticodon (the tRNA on the left) in Figure 15.11 were mutated from G to U. Give all codons with which the new, mutated anticodon could pair.
Question 15.29
Which of the following amino acid changes could result from a mutation that changed a single base? For each change that could result from the alteration of a single base, determine which position of the codon (first, second, or third nucleotide) in the mRNA must be altered for the change to result.
- a. Leu → Gln
- b. Phe → Ser
- c. Phe → Ile
- d. Pro → Ala
- e. Asn → Lys
- f. Ile → Asn
Section 15.3
Question 15.30
Arrange the following components of translation in the approximate order in which they would appear or be used in prokaryotic protein synthesis:
70S initiation complex
30S initiation complex
release factor 1
elongation factor G
initiation factor 3
elongation factor Tu
fMet-tRNAfMet
Question 15.31
Examine Figure 15.14 of a tRNA. What do you think would be the potential effect of a mutation in the part of the tRNA gene that encodes: (a) the acceptor stem; (b) the anticodon; (c) one of red-colored nucleotides?
Question 15.32
The following diagram illustrates a step in the process of translation. Sketch the diagram and identify the following elements on it.
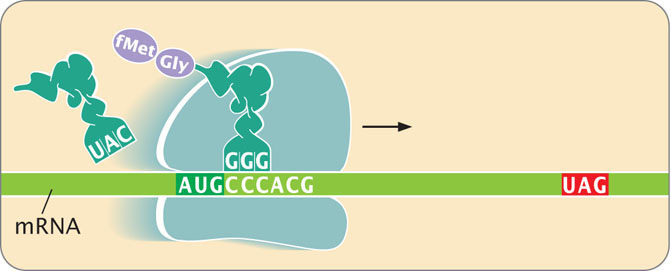
- a. 5′ and 3′ ends of the mRNA
- b. A, P, and E sites
- c. Start codon
- d. Stop codon
- e. Amino and carboxyl ends of the newly synthesized polypeptide chain
- f. Approximate location of the next peptide bond that will be formed
- g. Place on the ribosome where release factor 1 will bind
440
Question 15.33
Refer to the diagram in Problem 32 to answer the following questions.
- a. What will be the anticodon of the next tRNA added to the A site of the ribosome?
- b. What will be the next amino acid added to the growing polypeptide chain?
Question 15.34
A synthetic mRNA added to a cell-free protein-synthesizing system produces a peptide with the following amino acid sequence: Met-Pro-Ile-Ser-Ala. What would be the effect on translation if the following components were omitted from the cell-free protein-synthesizing system? What, if any, type of protein would be produced? Explain your reasoning.
- a. Initiation factor 3
- b. Initiation factor 2
- c. Elongation factor Tu
- d. Elongation factor G
- e. Release factors RF-1, RF-2, and RF-3
- f. ATP
- g. GTP
Question 15.35
For each of the following sequences, place a check mark in the appropriate space to indicate the process most immediately affected by deleting the sequence. Choose only one process for each sequence (i.e., one check mark per sequence).
| Process most immediately affected by deletion | ||||
|---|---|---|---|---|
| Sequence deleted | Replication | Transcription | RNA processing | Translation |
| a. ori site | _______ | _______ | _______ | _______ |
| b. 3′ splice-site consensus | _______ | _______ | _______ | _______ |
| c. poly(A) tail | _______ | _______ | _______ | _______ |
| d. terminator | _______ | _______ | _______ | _______ |
| e. start codon | _______ | _______ | _______ | _______ |
| f. −10 consensus | _______ | _______ | _______ | _______ |
| g. Shine–Dalgarno | _______ | _______ | _______ | _______ |
Question 15.36
 MicroRNAs are small RNA molecules that bind to the 3′ end of mRNAs and suppress translation (see Chapter 14). How miRNAs suppress translation is still being investigated. Some eukaryotic mRNAs have internal ribosome-binding sites downstream of the 5 cap, where ribosomes normally bind. In one investigation, miRNAs did not suppress the translation of ribosomes that attach to internal ribosome-binding sites (R. S. Pillai et al. 2005. Science 309:1573–1576). What does this finding suggest about how miRNAs suppress translation?
MicroRNAs are small RNA molecules that bind to the 3′ end of mRNAs and suppress translation (see Chapter 14). How miRNAs suppress translation is still being investigated. Some eukaryotic mRNAs have internal ribosome-binding sites downstream of the 5 cap, where ribosomes normally bind. In one investigation, miRNAs did not suppress the translation of ribosomes that attach to internal ribosome-binding sites (R. S. Pillai et al. 2005. Science 309:1573–1576). What does this finding suggest about how miRNAs suppress translation?
Question 15.37
Give the amino acid sequence of the protein encoded by the mRNA in Figure 15.21.
Section 15.4
Question 15.38
 Mutations that introduce stop codons cause a number of genetic diseases. For example, from 2% to 5% of the people who have cystic fibrosis possess a mutation that causes a premature stop codon in the gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR). This premature stop codon produces a truncated form of CFTR that is nonfunctional and results in the symptoms of cystic fibrosis. One possible way to treat people with genetic diseases caused by these types of mutations is to trick the ribosome into reading through the stop codon, inserting an amino acid into its place. Although the protein produced may have one altered amino acid, it is more likely to be at least partly functional than is the truncated protein produced when the ribosome stalls at the stop codon. Indeed, geneticists have conducted clinical trials on people with cystic fibrosis with the use of a drug called PTC124, which interferes with the ribosome’s ability to correctly read stop codons (C. Ainsworth. 2005. Nature 438:726–728). On the basis of what you know about the mechanism of nonsense-mediated mRNA decay (NMD), would you expect NMD to be a problem with this type of treatment? Why or why not?
Mutations that introduce stop codons cause a number of genetic diseases. For example, from 2% to 5% of the people who have cystic fibrosis possess a mutation that causes a premature stop codon in the gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR). This premature stop codon produces a truncated form of CFTR that is nonfunctional and results in the symptoms of cystic fibrosis. One possible way to treat people with genetic diseases caused by these types of mutations is to trick the ribosome into reading through the stop codon, inserting an amino acid into its place. Although the protein produced may have one altered amino acid, it is more likely to be at least partly functional than is the truncated protein produced when the ribosome stalls at the stop codon. Indeed, geneticists have conducted clinical trials on people with cystic fibrosis with the use of a drug called PTC124, which interferes with the ribosome’s ability to correctly read stop codons (C. Ainsworth. 2005. Nature 438:726–728). On the basis of what you know about the mechanism of nonsense-mediated mRNA decay (NMD), would you expect NMD to be a problem with this type of treatment? Why or why not?

441