15.4 Additional Properties of RNA and Ribosomes Affect Protein Synthesis
Now that we have considered in some detail the process of translation, we will examine some additional aspects of protein synthesis and the protein-synthesis machinery.
The Three-Dimensional Structure of the Ribosome
The central role of the ribosome in protein synthesis was recognized in the 1950s, and numerous aspects of its structure have been studied since then. Nevertheless, many aspects remained a mystery until detailed, three-dimensional reconstructions were completed. In 2009, the Nobel Prize in chemistry was awarded to Venkatraman Ramakrishnan, Thomas Steitz, and Ada Yonath for their research on the molecular structure of the ribosome.
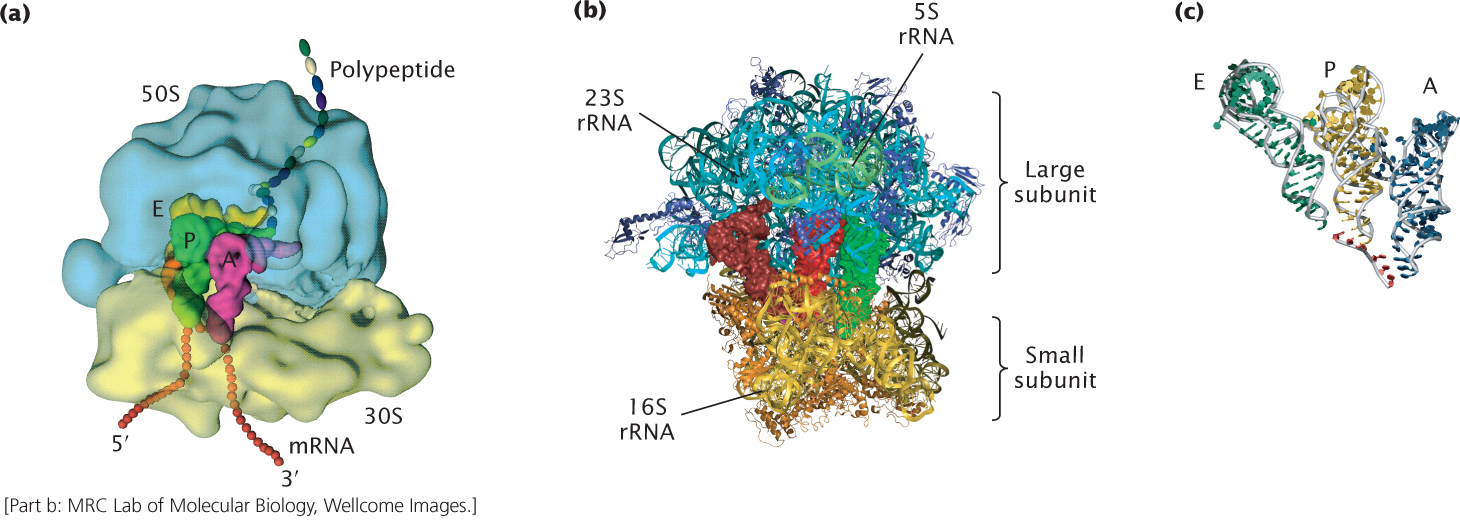
Figure 15.22a shows a model of the E. coli ribosome at low resolution. A high-resolution image of the bacterial ribosome as determined by X-ray crystallography is represented in the model depicted in Figure 15.22b. The mRNA is bound to the small subunit of the ribosome, and the tRNAs are located in the A, P, and E sites (Figure 15.22c) that bridge the small and large subunits (see Figure 15.22a). Initiation factors 1 and 3 bind to sites on the outside of the small subunit of the ribosome. EF-Tu, EF-G, and other factors complexed with GTP interact with a factor-binding center. High-resolution crystallographic images provide information indicating that a decoding center resides in the small subunit of the ribosome (the decoding center cannot be seen in Figure 15.22b). This center senses the fit between the codon on the mRNA and the anticodon on the incoming charged tRNA. Only tRNAs with the correct anticodon are bound tightly by the ribosome. The structural analyses also indicate that the large subunit of the ribosome contains the peptidyl transferase center, where peptide-bond formation takes place. There are no ribosomal proteins in the peptidyl transferase center; peptide-bond formation is carried out by the RNA molecule. These analyses also reveal that a tunnel connects the site of peptide-bond formation with the back of the ribosome; the growing polypeptide chain passes through this tunnel to the outside of the ribosome. The tunnel can accommodate about 35 amino acids of the growing polypeptide chain.
431
Polyribosomes
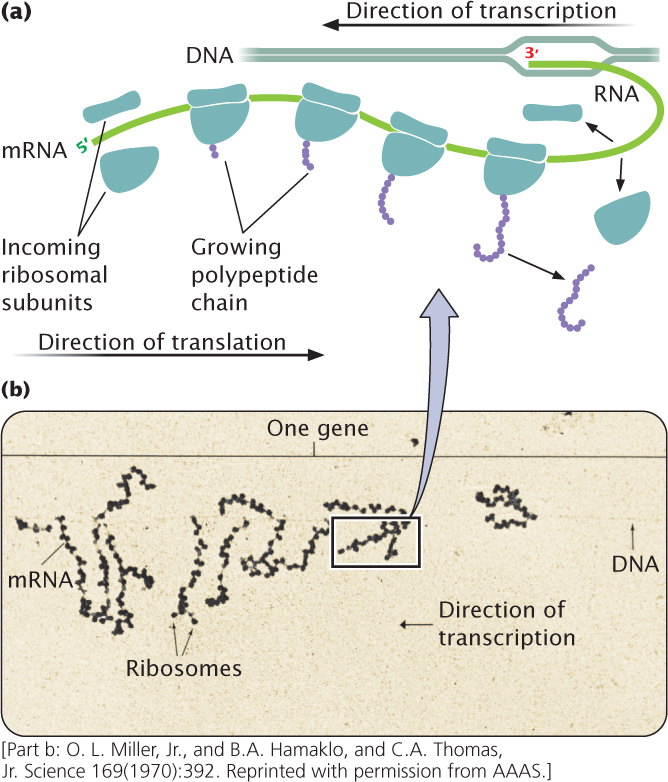
In both prokaryotic and eukaryotic cells, mRNA molecules are translated simultaneously by multiple ribosomes (Figure 15.23). The resulting structure—an mRNA with several ribosomes attached—is called a polyribosome (or often just a polysome). Each ribosome successively attaches to the ribosome-binding site at the 5′ end of the mRNA and moves toward the 3′ end; the polypeptide associated with each ribosome becomes progressively longer as the ribosome moves along the mRNA. In prokaryotic cells, transcription and translation are simultaneous; multiple ribosomes may be attached to the 5′ end of the mRNA while transcription is still taking place at the 3′ end, as shown in Figure 15.23. In eukaryotes, transcription and translation are separated in time and space, with transcription taking place in the nucleus and translation taking place in the cytoplasm.
CONCEPTS
In both prokaryotic and eukaryotic cells, multiple ribosomes may be attached to a single mRNA, generating a structure called a polyribosome.
 CONCEPT CHECK 9
CONCEPT CHECK 9
In a polyribosome, the polypeptides associated with which ribosomes will be the longest?
- Those at the 5′ end of mRNA.
- Those at the 3′ end of mRNA.
- Those in the middle of mRNA.
- All polypeptides will be the same length.
Messenger RNA Surveillance
The accurate transfer of genetic information from one generation to the next and from genotype to phenotype is critical for the proper development and functioning of an organism. Consequently, cells have evolved a number of quality-control mechanisms to ensure the accuracy of information transfer. Protein synthesis is no exception: several mechanisms, collectively termed mRNA surveillance, exist to detect and deal with errors in mRNAs that may create problems in the course of translation. These mechanisms keep the cell from wasting resources translating aberrant mRNAs and prevent the production of truncated proteins, which may be toxic to the cell.
432
Nonsense-Mediated mRNA Decay
A common mutation is one in which a codon that specifies an amino acid is altered to become a termination codon (called a nonsense mutation, see Chapter 18). A nonsense mutation does not affect transcription, but translation ends prematurely when the termination codon is encountered. The resulting protein is truncated and often nonfunctional. A common way that nonsense mutations arise in eukaryotic cells is when one or more of the exons is skipped or improperly spliced. Improper splicing leads to the deletion or addition of nucleotides in the mRNA, which alters the reading frame and often introduces premature termination codons.
To prevent the synthesis of aberrant proteins resulting from nonsense mutations, eukaryotic cells have evolved a mechanism called nonsense-mediated mRNA decay (NMD), which results in the rapid elimination of mRNA containing premature termination codons. The mechanism responsible for nonsense-mediated mRNA decay is still poorly understood. In mammals, it appears to entail proteins that bind to the exon–exon junctions. These exon–junction proteins may interact with enzymes that degrade the mRNA. One possibility is that the first ribosome to translate the mRNA (in the pioneer round of translation) removes the exon–junction proteins, thus protecting the mRNA from degradation. However, when the ribosome encounters a premature termination codon, the ribosome does not traverse the entire mRNA, and some of the exon–junction proteins are not removed, resulting in nonsense-mediated mRNA decay.  TRY PROBLEM 38
TRY PROBLEM 38
Stalled Ribosomes and Nonstop mRNAs
A problem that occasionally arises in translation is when a ribosome stalls on the mRNA before translation is terminated. This situation can arise when a mutation in the DNA changes a termination codon into a codon that specifies an amino acid. It can also arise when transcription terminates prematurely, producing a truncated mRNA lacking a termination codon. In these cases, the ribosome reaches the end of the mRNA without encountering a termination codon and stalls, still attached to the mRNA. Attachment to the mRNA prevents the ribosome from being recycled for use on other mRNAs and, if such occurrences are frequent, the result is a shortage of ribosomes that diminishes overall levels of protein synthesis.
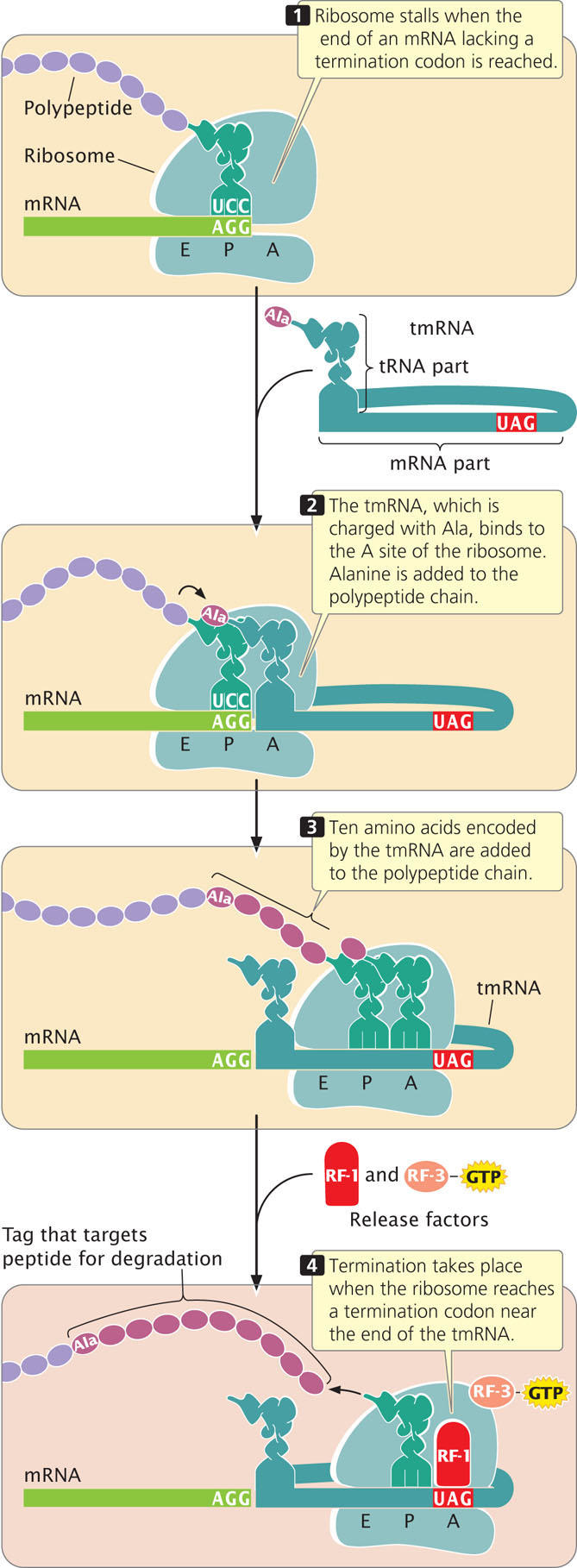
Bacteria have evolved a kind of molecular tow truck called transfer–messenger RNA (tmRNA) that removes stalled ribosomes. This RNA molecule has properties of both tRNA and mRNA; its tRNA component is normally charged with the amino acid alanine. When a ribosome becomes stalled on an mRNA, EF-Tu delivers the tmRNA to the ribosome’s A site, where tmRNA acts as surrogate tRNA (Figure 15.24). A peptide bond is created between the amino acid in the P site of the ribosome and alanine (attached to tmRNA) now in the A site, transferring the polypeptide chain to the tmRNA and releasing the tRNA in the P site.
433
The ribosome then resumes translation, switching from the original, aberrant mRNA to the mRNA part of tmRNA. Translation adds 10 amino acids encoded by the tmRNA, and then a termination codon is reached at the 3′ end of the tmRNA, which terminates translation and releases the ribosome. The added amino acids are a special tag that targets the incomplete polypeptide chain for degradation. Some evidence suggests that the tmRNA also targets the aberrant mRNA for degradation. How stalled ribosomes are recognized by the tmRNA is not clear, but this method is efficient at recycling stalled ribosomes and eliminating abnormal proteins that result from truncated transcription.
Eukaryotes have evolved a different mechanism to deal with mRNAs that are missing termination codons. Instead of restarting the stalled ribosome and degrading the abnormal protein that results, eukaryotic cells use a mechanism called nonstop mRNA decay, which results in the rapid degradation of abnormal mRNA. In this mechanism, the codonfree A site of the stalled ribosome is recognized by a special protein that binds to the A site and recruits other proteins, which then degrade the mRNA from its 3′ end.
No-Go Decay
Another mRNA surveillance system found in eukaryotes is no-go decay (NGD), which helps remove stalled ribosomes resulting from secondary structures in the mRNA, chemical damage to the mRNA, premature stop codons, and ribosomal defects. A series of proteins bring about termination, recycling of the ribosomes, and degradation of the mRNA.
CONCEPTS
Cells possess mRNA surveillance mechanisms to detect and eliminate mRNA molecules containing errors that create problems in the course of translation.
Folding and Posttranslational Modifications of Proteins
The functions of many proteins critically depend on the proper folding of the polypeptide chain; some proteins spontaneously fold into their correct shapes, but, for others, correct folding may initially require the participation of other molecules called molecular chaperones. Some molecular chaperones are associated with the ribosome and fold newly synthesized polypeptide chains as they emerge from the ribosome tunnel, in which case protein folding takes place during ongoing translation.
Many proteins must be modified after translation to become active. Proteins in both prokaryotic and eukaryotic cells often undergo alterations following translation, which are termed posttranslational modifications. A number of different types of modifications are possible. Some proteins are synthesized as larger precursor proteins and must be cleaved and trimmed by enzymes before the proteins can become functional. As mentioned earlier, the formyl group or the entire methionine residue may be removed from the amino end of a protein. Some proteins require the attachment of carbohydrates for activation. Amino acids within a protein may be modified: phosphates, carboxyl groups, and methyl groups are added to some amino acids. In eukaryotic cells, the amino end of a protein is often acetylated after translation.
A common posttranslational modification in eukaryotes is the attachment of a protein called ubiquitin, which targets the protein for degradation. Another modification of some proteins is the removal of 15 to 30 amino acids, called the signal sequence, at the amino end of the protein. The signal sequence helps direct a protein to a specific location within the cell, after which the sequence is removed by special enzymes.
CONCEPTS
Many proteins undergo posttranslational modifications after their synthesis.
Translation and Antibiotics
Antibiotics are drugs that kill microorganisms. To make an effective antibiotic, not just any poison will do: the trick is to kill the bacteria without harming the patient.
Translation is frequently the target of antibiotics because it is essential to all living organisms and differs significantly between bacterial and eukaryotic cells. A number of antibiotics bind selectively to bacterial ribosomes and inhibit various steps in translation, but they do not affect eukaryotic ribosomes. Tetracyclines, for instance, are a class of antibiotics that bind to the A site of a bacterial ribosome and block the entry of charged tRNAs, yet they have no effect on eukaryotic ribosomes. Neomycin binds to the ribosome near the A site and induces translational errors, probably by causing mistakes in the binding of charged tRNAs to the A site. Chloramphenicol binds to the large subunit of the ribosome and blocks peptide-bond formation. Streptomycin binds to the small subunit of the ribosome and inhibits initiation, and erythromycin blocks translocation. Although chloramphenicol and streptomycin are potent inhibitors of translation in bacteria, they do not inhibit translation in archaea.
The three-dimensional structure of puromycin resembles the 3′ end of a charged tRNA, permitting puromycin to enter the A site of a ribosome efficiently and inhibit the entry of tRNAs. A peptide bond can form between the puromycin molecule in the A site and an amino acid on the tRNA in the P site of the ribosome, but puromycin cannot bind to the P site and translocation does not take place, blocking further elongation of the protein. Because tRNA structure is similar in all organisms, puromycin inhibits translation in both bacterial and eukaryotic cells; consequently, puromycin kills eukaryotic cells along with bacteria and is sometimes used in cancer therapy to destroy tumor cells.
Many antibiotics act by blocking specific steps in translation, and different antibiotics block different steps in protein synthesis, such as initiation or elongation. Because of this specificity, antibiotics are frequently used to study the process of protein synthesis.
434