16.4 RNA Molecules Control the Expression of Some Bacterial Genes
All the regulators of gene expression that we have considered so far have been proteins. Several examples of RNA regulators also have been discovered.
Antisense RNA
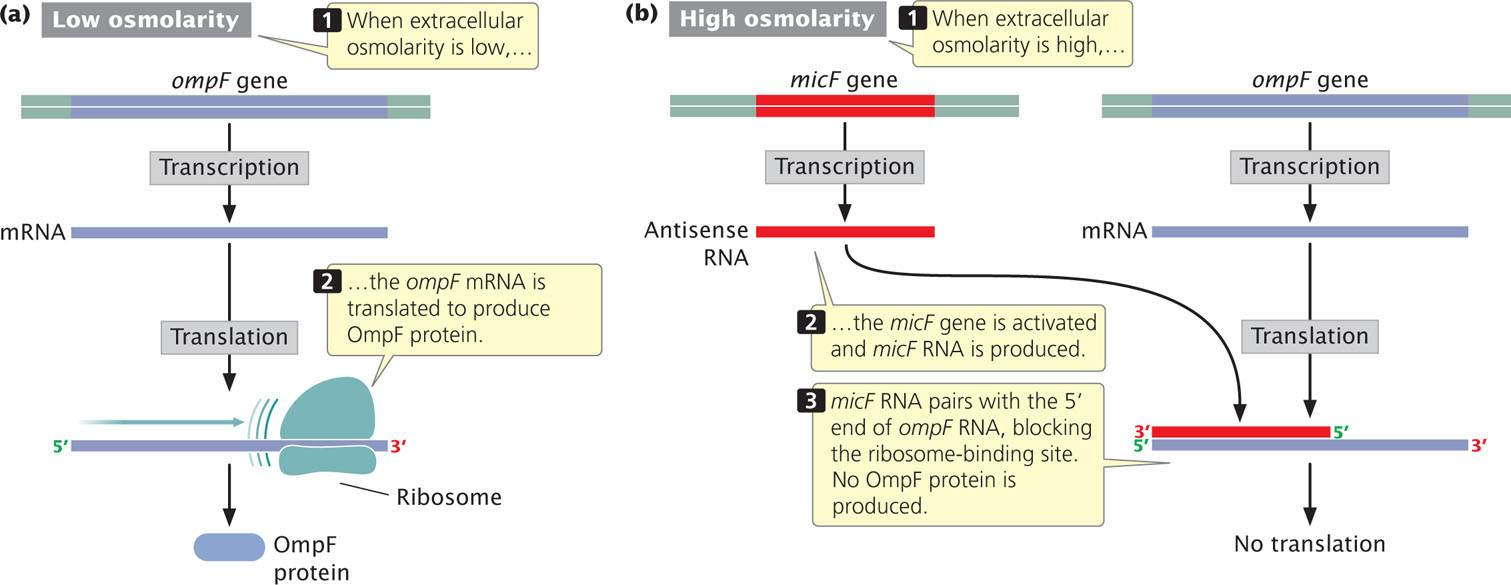
Some RNA molecules are complementary to particular sequences on mRNAs and are called antisense RNA. They control gene expression by binding to sequences on mRNA and inhibiting translation. Translational control by antisense RNA is seen in the regulation of the ompF gene of E. coli (Figure 16.18a). This gene encodes an outer-membrane protein that functions as a channel for the passive diffusion of small polar molecules, such as water and ions, across the cell membrane. Under most conditions, the ompF gene is transcribed and translated and the OmpF protein is synthesized. However, when the osmolarity of the medium increases, the cell depresses the production of OmpF protein to help maintain cellular osmolarity. A regulator gene named micF—for mRNA-interfering complementary RNA—is activated and micF RNA is produced (Figure 16.18b). The micF RNA, an antisense RNA, binds to a complementary sequence in the 5′ UTR of the ompF mRNA and inhibits the binding of the ribosome. This inhibition reduces the amount of translation (see Figure 16.18b), which results in fewer OmpF proteins in the outer membrane and thus reduces the detrimental movement of substances across the membrane owing to the changes in osmolarity. Recent research has found many antisense RNA molecules in a wide range of bacterial species.  TRY PROBLEM 30
TRY PROBLEM 30
Riboswitches
We have seen that operons of bacteria contain DNA sequences (promoters and operator sites) where the binding of small molecules induces or represses transcription. Some mRNA molecules contain regulatory sequences called riboswitches, where molecules can bind and affect gene expression by influencing the formation of secondary structures in the mRNA (Figure 16.19). Riboswitches were first discovered in 2002 and now appear to be common in bacteria, regulating about 4% of all bacterial genes. They are also present in archaea, fungi, and plants.
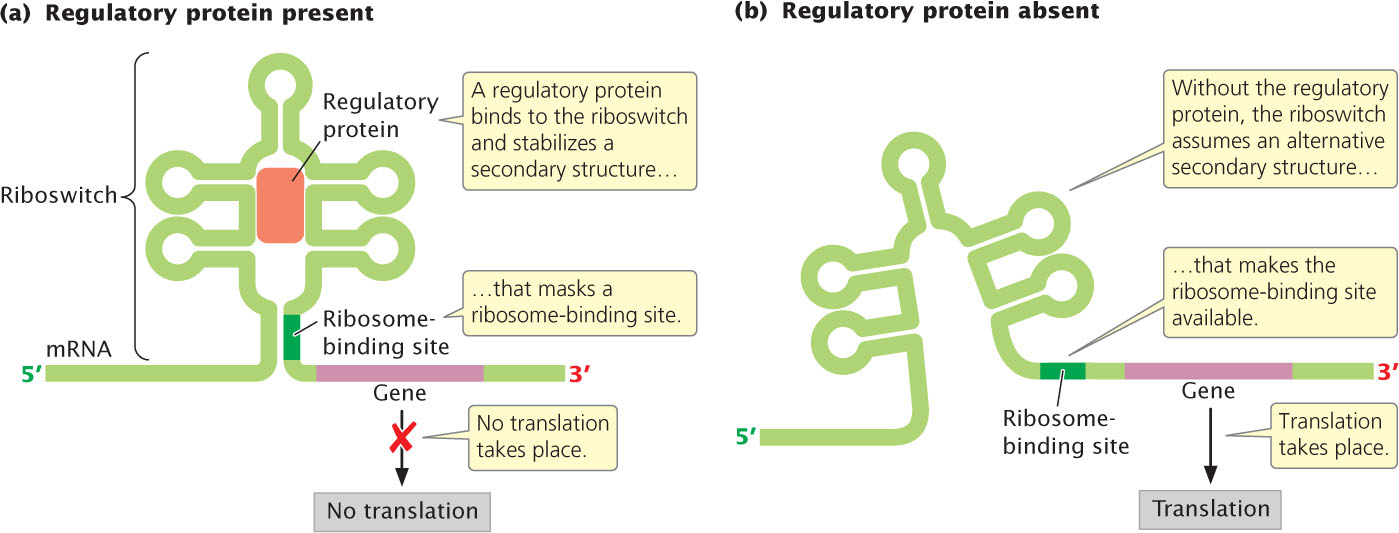
Riboswitches are typically found in the 5′ UTR of the mRNA and fold into compact RNA secondary structures with a base stem and several branching hairpins. In some cases, a small regulatory molecule binds to the riboswitch and stabilizes a terminator, which causes premature termination of transcription. In other cases, the binding of a regulatory molecule stabilizes a secondary structure that masks the ribosome-binding site, preventing the initiation of translation. When not bound by the regulatory molecule, the riboswitch assumes an alternative structure that eliminates the premature terminator or makes the ribosome-binding site available.
465
A Riboswitch Controls the Synthesis of Vitamin B12
An example of a riboswitch is seen in bacterial genes that encode enzymes having roles in the synthesis of vitamin B12. The genes for these enzymes are transcribed into an mRNA molecule that has a riboswitch. When the activated form of vitamin B12—called coenzyme B12—is present, it binds to the riboswitch and the mRNA folds into a secondary structure that obstructs the ribosome-binding site. Consequently, no translation of the mRNA takes place. In the absence of coenzyme B12, the mRNA assumes a different secondary structure. This secondary structure does not obstruct the ribosome-binding site, and so translation is initiated, the enzymes are synthesized, and more vitamin B12 is produced. For some riboswitches, the regulatory molecule acts as a repressor (as just described) by inhibiting transcription or translation; for others, the regulatory molecule acts as an inducer by causing the formation of a secondary structure that allows transcription or translation to take place.
466
RNA-Mediated Repression Through Ribozymes
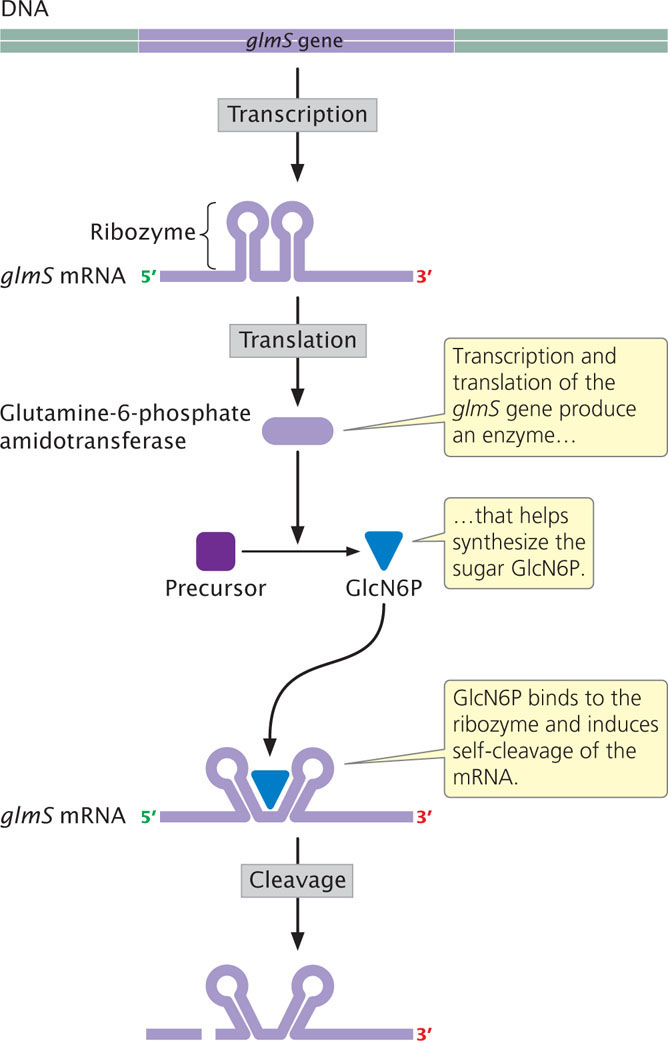
Another type of gene control is carried out by mRNA molecules called ribozymes, which possess catalytic activity (see Chapter 14). Termed RNA-mediated repression, this type of control has been demonstrated in the glmS gene of the bacterium Bacillus subtilis. Transcription of this gene produces an mRNA molecule that encodes the enzyme glutamine-fructose-6-phosphate amidotransferase (Figure 16.20), which helps synthesize a small sugar called glucosamine-6-phosphate (GlcN6P). Within the 5′ UTR of the glmS mRNA are about 75 nucleotides that act as a ribozyme. When GlcN6P is absent, the glmS gene is transcribed and translated to produce the enzyme, which synthesizes more GlcN6P. However, when sufficient GlcN6P is present, it binds to the ribozyme part of the mRNA, which then induces self-cleavage of the mRNA—the ribozyme breaks the sugar-phosphate backbone of the RNA. This cleavage then prevents translation of the mRNA.
CONCEPTS
Antisense RNA is complementary to other RNA or DNA sequences. In bacterial cells, it can inhibit translation by binding to sequences in the 5′ UTR of mRNA and preventing the attachment of the ribosome. Riboswitches are sequences in mRNA molecules that bind regulatory molecules and induce changes in the secondary structure of the mRNA that affect gene expression. In RNA-mediated repression, a ribozyme sequence on the mRNA induces self-cleavage and degradation of the mRNA when bound by a regulatory molecule.