Application Questions and Problems
Introduction
Question 7.13
The introduction to this chapter described the search for genes that determine pattern baldness in humans. In 1916, Dorothy Osborn suggested that pattern baldness is a sex-influenced trait (see Chapter 5) that is dominant in males and recessive in females. More research suggested that pattern baldness is an X-linked recessive trait. Would you expect to see independent assortment between genetic markers on the X chromosome and pattern baldness if (a) pattern baldness is sex-influenced and (b) if pattern baldness is X-linked recessive? Explain your answer.
Section 7.2
Question 7.14
In the snail Cepaea nemoralis, an autosomal allele causing a banded shell (BB) is recessive to the allele for an unbanded shell (BO). Genes at a different locus determine the background color of the shell; here, yellow (CY) is recessive to brown (CBw). A banded, yellow snail is crossed with a homozygous brown, unbanded snail. The F1 are then crossed with banded, yellow snails (a testcross).
- a. What will the results of the testcross be if the loci that control banding and color are linked with no crossing over?
- b. What will the results of the testcross be if the loci assort independently?
 [Otto Hahn/Getty Images.]
[Otto Hahn/Getty Images.] - c. What will the results of the testcross be if the loci are linked and 20 m.u. apart?
Question 7.15
In silkmoths (Bombyx mori), red eyes (re) and white-banded wing (wb) are encoded by two mutant alleles that are recessive to those that produce wild-type traits (re+ and wb+); these two genes are on the same chromosome. A moth homozygous for red eyes and white-banded wings is crossed with a moth homozygous for the wild-type traits. The F1 have normal eyes and normal wings. The F1 are crossed with moths that have red eyes and white-banded wings in a testcross. The progeny of this testcross are:
| wild-type eyes, wild-type wings | 418 |
| red eyes, wild-type wings | 19 |
| wild-type eyes, white-banded wings | 16 |
| red eyes, white-banded wings | 426 |
- a. What phenotypic proportions would be expected if the genes for red eyes and for white-banded wings were located on different chromosomes?
- b. What is the percent recombination between the genes for red eyes and those for white-banded wings?
Question 7.16
A geneticist discovers a new mutation in Drosophila melanogaster that causes the flies to shake and quiver. She calls this mutation spastic (sps) and determines that it is due to an autosomal recessive gene. She wants to determine whether the gene encoding spastic is linked to the recessive gene for vestigial wings (vg). She crosses a fly homozygous for spastic and vestigial traits with a fly homozygous for the wild-type traits and then uses the resulting F1 females in a testcross. She obtains the following flies from this testcross.
| vg+ | sps+ | 230 |
| vg | sps | 224 |
| vg | sps+ | 97 |
| vg+ | sps | 99 |
| Total | 650 |
Are the genes that cause vestigial wings and the spastic mutation linked? Do a chi-square test of independence to determine whether the genes have assorted independently.
Question 7.17
In cucumbers, heart-shaped leaves (hl) are recessive to normal leaves (Hl) and having numerous fruit spines (ns) is recessive to having few fruit spines (Ns). The genes for leaf shape and for number of spines are located on the same chromosome; findings from mapping experiments indicate that they are 32.6 m.u. apart. A cucumber plant having heart-shaped leaves and numerous spines is crossed with a plant that is homozygous for normal leaves and few spines. The F1 are crossed with plants that have heart-shaped leaves and numerous spines. What phenotypes and phenotypic proportions are expected in the progeny of this cross?
Question 7.18
In tomatoes, tall (D) is dominant over dwarf (d) and smooth fruit (P) is dominant over pubescent fruit (p), which is covered with fine hairs. A farmer has two tall and smooth tomato plants, which we will call plant A and plant B. The farmer crosses plants A and B with the same dwarf and pubescent plant and obtains the following numbers of progeny:
| Progeny of | ||
|---|---|---|
| Plant A | Plant B | |
| Dd Pp | 122 | 2 |
| Dd pp | 6 | 82 |
| dd Pp | 4 | 82 |
| dd pp | 124 | 4 |
- a. What are the genotypes of plant A and plant B?
- b. Are the loci that determine the height of the plant and pubescence linked? If so, what is the percent recombination between them?
- c. Explain why different proportions of progeny are produced when plant A and plant B are crossed with the same dwarf pubescent plant.
Question 7.19
Alleles A and a are at a locus on the same chromosome as is a locus with alleles B and b. Aa Bb is crossed with aa bb and the following progeny are produced:
| Aa Bb | 5 |
| Aa bb | 45 |
| aa Bb | 45 |
| aa bb | 5 |
What conclusion can be made about the arrangement of the genes on the chromosome in the Aa Bb parent?
Question 7.20
 Daniel McDonald and Nancy Peer determined that eyespot (a clear spot in the center of the eye) in flour beetles is caused by an X-linked gene (es) that is recessive to the allele for the absence of eyespot (es+). They conducted a series of crosses to determine the distance between the gene for eyespot and a dominant X-linked gene for striped (St), which causes white stripes on females and acts as a recessive lethal (is lethal when homozygous in females or hemizygous in males). The following cross was carried out (D. J. McDonald and N. J. Peer. 1961. Journal of Heredity 52:261–264).
Daniel McDonald and Nancy Peer determined that eyespot (a clear spot in the center of the eye) in flour beetles is caused by an X-linked gene (es) that is recessive to the allele for the absence of eyespot (es+). They conducted a series of crosses to determine the distance between the gene for eyespot and a dominant X-linked gene for striped (St), which causes white stripes on females and acts as a recessive lethal (is lethal when homozygous in females or hemizygous in males). The following cross was carried out (D. J. McDonald and N. J. Peer. 1961. Journal of Heredity 52:261–264).

- a. Which progeny are the recombinants and which progeny are the nonrecombinants?
- b. Calculate the recombination frequency between es and St.
- c. Are some potential genotypes missing among the progeny of the cross? If so, which ones and why?
Question 7.21
Recombination rates between three loci in corn are shown here.
| Loci | Recombination rate |
|---|---|
| R and W2 | 17% |
| R and L2 | 35% |
| W2 and L2 | 18% |
What is the order of the genes on the chromosome?
Question 7.22
In tomatoes, dwarf (d) is recessive to tall (D) and opaque (light-green) leaves (op) are recessive to green leaves (Op). The loci that determine height and leaf color are linked and separated by a distance of 7 m.u. For each of the following crosses, determine the phenotypes and proportions of progeny produced.
- a.
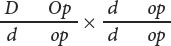
- b.
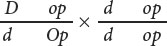
- c.
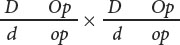
- d.
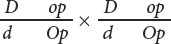
Question 7.23
In German cockroaches, bulging eyes (bu) are recessive to normal eyes (bu+) and curved wings (cv) are recessive to straight wings (cv+). Both traits are encoded by autosomal genes that are linked. A cockroach has genotype bu+bu cv+cv, and the genes are in replusion. Which of the following sets of genes will be found in the most-common gametes produced by this cockroach?
- a. bu+ cv+
- b. bu cv
- c. bu+ bu
- d. cv+ cv
- e. bu cv+
Explain your answer.
Question 7.24
In Drosophila melanogaster, ebony body (e) and rough eyes (ro) are encoded by autosomal recessive genes found on chromosome 3; they are separated by 20 m.u. The gene that encodes forked bristles (f) is X-linked recessive and assorts independently of e and ro. Give the phenotypes of progeny and their expected proportions when a female of each of the following genotypes is test-crossed with a male.
- a.
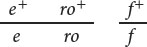
- b.
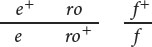
Question 7.25
 Honeybees have haplodiploid sex determination: females are diploid, developing from fertilized eggs, whereas males are haploid, developing from unfertilized eggs. Otto Mackensen studied linkage relations among eight mutations in honeybees (O. Mackensen. 1958. Journal of Heredity 49:99–102). The following table gives the results of two of MacKensen’s crosses including three recessive mutations: cd (cordovan body color), h (hairless), and ch (chartreuse eye color).
Honeybees have haplodiploid sex determination: females are diploid, developing from fertilized eggs, whereas males are haploid, developing from unfertilized eggs. Otto Mackensen studied linkage relations among eight mutations in honeybees (O. Mackensen. 1958. Journal of Heredity 49:99–102). The following table gives the results of two of MacKensen’s crosses including three recessive mutations: cd (cordovan body color), h (hairless), and ch (chartreuse eye color).
204
| Queen genotype | Phenotypes of drone (male) progeny |
|---|---|
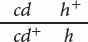
|
294 cordovan, 236 hairless, 262 cordovan and hairless, 289 wild type |
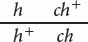
|
3131 hairless, 3064 chartreuse, 96 chartreuse and hairless, 132 wild type |
- a. Only the genotype of the queen is given. Why is the genotype of the male parent not needed for mapping these genes? Would the genotype of the male parent be required if we examined female progeny instead of male progeny?
- b. Determine the nonrecombinant and recombinant progeny for each cross and calculate the map distances between cd, h, and ch. Draw a linkage map illustrating the linkage arrangements among these three genes.
Question 7.26
Perform a chi-square test of independence on the data provided in Figure 7.2 to determine if the genes for flower color and pollen shape in sweet peas are assorting independently. Give the chi-square value, degrees of freedom, and associated probability. What conclusion would you make about the independent assortment of these genes?
Question 7.27
A series of two-point crosses were carried out among seven loci (a, b, c, d, e, f, and g), producing the following recombination frequencies. Map the seven loci, showing their linkage groups, the order of the loci in each linkage group, and the distances between the loci of each group.
| Loci | Percent recombination | Loci | Percent recombination |
|---|---|---|---|
| a and b | 50 | c and d | 50 |
| a and c | 50 | c and e | 26 |
| a and d | 12 | c and f | 50 |
| a and e | 50 | c and g | 50 |
| a and f | 50 | d and e | 50 |
| a and g | 4 | d and f | 50 |
| b and c | 10 | d and g | 8 |
| b and d | 50 | e and f | 50 |
| b and e | 18 | e and g | 50 |
| b and f | 50 | f and g | 50 |
| b and g | 50 |
Question 7.28
 R. W. Allard and W. M. Clement determined recombination rates for a series of genes in lima beans (R. W. Allard and W. M. Clement. 1959. Journal of Heredity 50:63–67). The following table lists paired recombination rates for eight of the loci (D, Wl, R, S, L1, Ms, C, and G) that they mapped. On the basis of these data, draw a series of genetic maps for the different linkage groups of the genes, indicating the distances between the genes. Keep in mind that these rates are estimates of the true recombination rates and that some error is associated with each estimate. An asterisk beside a recombination frequency indicates that the recombination frequency is significantly different from 50%.
R. W. Allard and W. M. Clement determined recombination rates for a series of genes in lima beans (R. W. Allard and W. M. Clement. 1959. Journal of Heredity 50:63–67). The following table lists paired recombination rates for eight of the loci (D, Wl, R, S, L1, Ms, C, and G) that they mapped. On the basis of these data, draw a series of genetic maps for the different linkage groups of the genes, indicating the distances between the genes. Keep in mind that these rates are estimates of the true recombination rates and that some error is associated with each estimate. An asterisk beside a recombination frequency indicates that the recombination frequency is significantly different from 50%.
| Recombination Rates (%) among Seven Loci in Lima Beans | |||||||
| Wl | R | S | L1 | Ms | C | G | |
| D | 2.1* | 39.3* | 52.4 | 48.1 | 53.1 | 51.4 | 49.8 |
| Wl | 38.0* | 47.3 | 47.7 | 48.8 | 50.3 | 50.4 | |
| R | 51.9 | 52.7 | 54.6 | 49.3 | 52.6 | ||
| S | 26.9* | 54.9 | 52.0 | 48.0 | |||
| Li | 48.2 | 45.3 | 50.4 | ||||
| Ms | 14.7* | 43.1 | |||||
| C | 52.0 | ||||||
| *Significantly different from 50%. | |||||||
Section 7.3
Question 7.29
 Raymond Popp studied linkage among genes for pink eye (p), shaker-1 (sh-1, which causes circling behavior, head tossing, and deafness), and hemoglobin (Hb) in mice (R. A. Popp. 1962. Journal of Heredity 53:73–80). He performed a series of testcrosses, in which mice heterozygous for pink eye, shaker-1, and hemoglobin 1 and 2 were crossed with mice that were homozygous for pink eye, shaker-1, and hemoglobin 2.
Raymond Popp studied linkage among genes for pink eye (p), shaker-1 (sh-1, which causes circling behavior, head tossing, and deafness), and hemoglobin (Hb) in mice (R. A. Popp. 1962. Journal of Heredity 53:73–80). He performed a series of testcrosses, in which mice heterozygous for pink eye, shaker-1, and hemoglobin 1 and 2 were crossed with mice that were homozygous for pink eye, shaker-1, and hemoglobin 2.
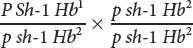
The following progeny were produced.

205
- a. Determine the order of these genes on the chromosome.
- b. Calculate the map distances between the genes.
- c. Determine the coefficient of coincidence and the interference among these genes.
Question 7.30
Waxy endosperm (wx), shrunken endosperm (sh), and yellow seedling (v) are encoded by three recessive genes in corn that are linked on chromosome 5. A corn plant homozygous for all three recessive alleles is crossed with a plant homozygous for all the dominant alleles. The resulting F1 are then crossed with a plant homozygous for the recessive alleles in a three-point testcross. The progeny of the testcross are:
| wx | sh | V | 87 |
| Wx | Sh | v | 94 |
| Wx | Sh | V | 3,479 |
| wx | sh | v | 3,478 |
| Wx | sh | V | 1,515 |
| wx | Sh | v | 1,531 |
| wx | Sh | V | 292 |
| Wx | sh | v | 280 |
| Total | 10,756 |
- a. Determine the order of these genes on the chromosome.
- b. Calculate the map distances between the genes.
- c. Determine the coefficient of coincidence and the interference among these genes.
Question 7.31
 Priscilla Lane and Margaret Green studied the linkage relations of three genes affecting coat color in mice: mahogany (mg), agouti (a), and ragged (Rg). They carried out a series of three-point crosses, mating mice that were heterozygous at all three loci with mice that were homozygous for the recessive alleles at these loci (P W. Lane and M. C. Green. 1960. Journal of Heredity 51:228–230). The following table lists the results of the testcrosses.
Priscilla Lane and Margaret Green studied the linkage relations of three genes affecting coat color in mice: mahogany (mg), agouti (a), and ragged (Rg). They carried out a series of three-point crosses, mating mice that were heterozygous at all three loci with mice that were homozygous for the recessive alleles at these loci (P W. Lane and M. C. Green. 1960. Journal of Heredity 51:228–230). The following table lists the results of the testcrosses.
| Phenotype | Number | ||
|---|---|---|---|
| a | Rg | + | 1 |
| + | + | mg | 1 |
| a | + | + | 15 |
| + | Rg | mg | 9 |
| + | + | + | 16 |
| a | Rg | mg | 36 |
| a | + | mg | 76 |
| + | Rg | + | 69 |
| Total | 213 | ||
| Note: + represents a wild-type allele. | |||
- a. Determine the order of the loci that encode mahogany, agouti, and ragged on the chromosome, the map distances between them, and the interference and coefficient of coincidence for these genes.
- b. Draw a picture of the two chromosomes in the triply heterozygous mice used in the testcrosses, indicating which of the alleles are present on each chromosome.
Question 7.32
Fine spines (s), smooth fruit (tu), and uniform fruit color (u) are three recessive traits in cucumbers, the genes of which are linked on the same chromosome. A cucumber plant heterozygous for all three traits is used in a testcross, and the following progeny are produced from this testcross:
| S | U | Tu | 2 |
| s | u | Tu | 70 |
| S | u | Tu | 21 |
| s | u | tu | 4 |
| S | U | tu | 82 |
| s | U | tu | 21 |
| s | U | Tu | 13 |
| S | u | tu | 17 |
| Total | 230 |
- a. Determine the order of these genes on the chromosome.
- b. Calculate the map distances between the genes.
- c. Determine the coefficient of coincidence and the interference among these genes.
- d. List the genes found on each chromosome in the parents used in the testcross.
Question 7.33
In Drosophila melanogaster, black body (b) is recessive to gray body (b+), purple eyes (pr) are recessive to red eyes (pr+), and vestigial wings (vg) are recessive to normal wings (vg+). The loci encoding these traits are linked, with the following map distances:
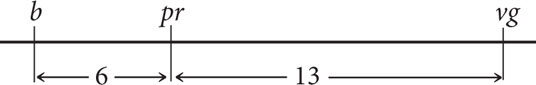
The interference among these genes is 0.5. A fly with a black body, purple eyes, and vestigial wings is crossed with a fly homozygous for a gray body, red eyes, and normal wings. The female progeny are then crossed with males that have a black body, purple eyes, and vestigial wings. If 1000 progeny are produced from this testcross, what will be the phenotypes and proportions of the progeny?
Question 7.34
Sepia eyes, spineless bristles, and striped thorax are three recessive mutations in Drosophila found on chromosome 3. A genetics student crosses a fly homozygous for sepia eyes, spineless bristles, and striped thorax with a fly homozygous for the wild-type traits—red eyes, normal bristles, and solid thorax. The female progeny are then test-crossed with males that have sepia eyes, spineless bristles, and striped thorax. Assume that the interference between these genes is 0.2 and that 400 progeny flies are produced from the test cross. Based on the map distances provided in Figure 7.15, predict the phenotypes and proportions of the progeny resulting from the test cross.
206
Question 7.35
Shown below are eight DNA sequences from different individuals.
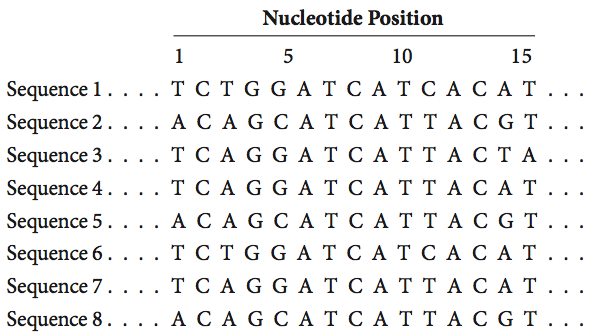
- a. Give the nucleotide positions of all single nucleotide polymorphisms (SNPs; nucleotide positions where individuals vary in which base is present) in these sequences.
- b. How many different haplotypes (sets of linked variants) are found in these eight sequences?
- c. Give the haplotype of each sequence by listing the specific bases at each variable position in that particular haplotype. (Hint: See Figure 20.8)
Question 7.36
A group of geneticists are interested in identifying genes that may play a role in susceptibility to asthma. They study the inheritance of genetic markers in a series of families that have two or more asthmatic children. They find an association between the presence or absence of asthma and a genetic marker on the short arm of chromosome 20 and calculate a lod score of 2 for this association. What does this lod score indicate about genes that may influence asthma?
Section 7.4
Question 7.37
A panel of cell lines was created from human–mouse somatic-cell fusions. Each line was examined for the presence of human chromosomes and for the production of an enzyme. The following results were obtained:
| Human chromosomes | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell line | Enzyme | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 17 | 22 |
| A | − | + | − | − | − | + | − | − | − | − | − | + | − |
| B | + | + | + | − | − | − | − | − | + | − | − | + | + |
| C | − | + | − | − | − | + | − | − | − | − | − | − | + |
| D | − | − | − | − | + | − | − | − | − | − | − | − | − |
| E | + | + | − | − | − | − | − | − | + | − | + | + | − |
On the basis of these results, which chromosome has the gene that encodes the enzyme?
Question 7.38
A panel of cell lines was created from human–mouse somatic-cell fusions. Each line was examined for the presence of human chromosomes and for the production of three enzymes. The following results were obtained.
| Cell line | Enzyme | Human chromosomes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 8 | 9 | 12 | 15 | 16 | 17 | 22 | X | ||
| A | + | − | + | − | − | + | − | + | + | − | − | + | |
| B | + | − | − | − | − | + | − | − | + | + | − | − | |
| C | − | + | + | + | − | − | − | − | − | + | − | + | |
| D | − | + | + | + | + | − | − | − | + | − | − | + | |
On the basis of these results, give the chromosome location of enzyme 1, enzyme 2, and enzyme 3.
Question 7.39
The locations of six deletions have been mapped to the Drosophila chromosome, as shown in the following deletion map. Recessive mutations a, b, c, d, e, and f are known to be located in the same region as the deletions, but the order of the mutations on the chromosome is not known.
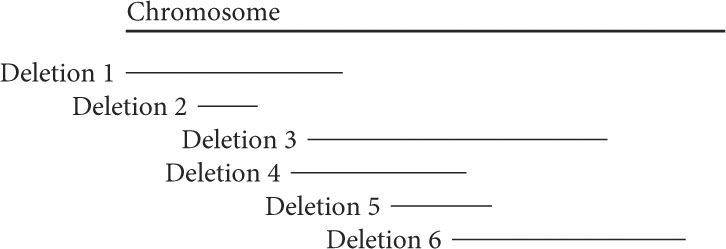
When flies homozygous for the recessive mutations are crossed with flies heterozygous for the deletions, the following results are obtained, in which “m” represents a mutant phenotype and a plus sign (+) represents the wild type. On the basis of these data, determine the relative order of the seven mutant genes on the chromosome:
| Mutations | ||||||
| Deletion | a | b | c | d | e | f |
| 1 | m | + | m | + | + | m |
| 2 | m | + | + | + | + | + |
| 3 | + | m | m | m | m | + |
| 4 | + | + | m | m | m | + |
| 5 | + | + | + | m | m | + |
| 6 | + | m | + | m | + | + |
207