Answers to Concept Checks
- 1. c
- 2. Repulsion
- 3. Genetic maps are based on rates of recombination; physical maps are based on physical distances.
- 4.
- 5. The c locus
- 6. b
WORKED PROBLEMS
Problem 1
In guinea pigs, white coat (w) is recessive to black coat (W) and wavy hair (v) is recessive to straight hair (V). A breeder crosses a guinea pig that is homozygous for white coat and wavy hair with a guinea pig that is black with straight hair. The F1 are then crossed with guinea pigs having white coats and wavy hair in a series of testcrosses. The following progeny are produced from these testcrosses:
197
| black, straight | 30 |
| black, wavy | 10 |
| white, straight | 12 |
| white, wavy | 31 |
| — | |
| Total | 83 |
- a. Are the genes that determine coat color and hair type assorting independently? Carry out chi-square tests to test your hypothesis.
- b. If the genes are not assorting independently, what is the recombination frequency between them?
Solution Strategy
What information is required in your answer to the problem?
- a. If the genes are assorting independently, along with a chi-square value, d.f. and P value to evaluate your hypothesis.
- b. If the genes are not assorting independently, the recombination frequency between them.
What information is provided to solve the problem?
 White coat is recessive to black coat and wavy hair is recessive to straight hair.
White coat is recessive to black coat and wavy hair is recessive to straight hair. The numbers of different types of progeny from a series of testcrosses.
The numbers of different types of progeny from a series of testcrosses.
For help with this problem, review:
Crossing Over with Linked Genes, Calculating Recombination Frequency and Testing for Independent Assortment in Section 7.2
Solution Steps
- a. Assuming independent assortment, outline the crosses conducted by the breeder:
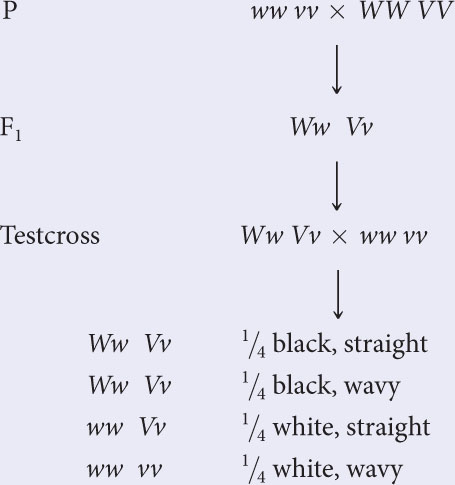
Recall: With independent assortment, we expect equal numbers of nonrecombinant and recombinant offspring.
Because a total of 83 progeny were produced in the testcrosses, we expect 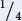 × 83 = 20.75 of each. The observed numbers of progeny from the testcross (30, 10, 12, 31) do not appear to fit the expected numbers (20.75, 20.75, 20.75, 20.75) well; so independent assortment may not have taken place.
× 83 = 20.75 of each. The observed numbers of progeny from the testcross (30, 10, 12, 31) do not appear to fit the expected numbers (20.75, 20.75, 20.75, 20.75) well; so independent assortment may not have taken place.
Hint: See Figure 7.10 for a review of how to carry out a chi-square test of independence.
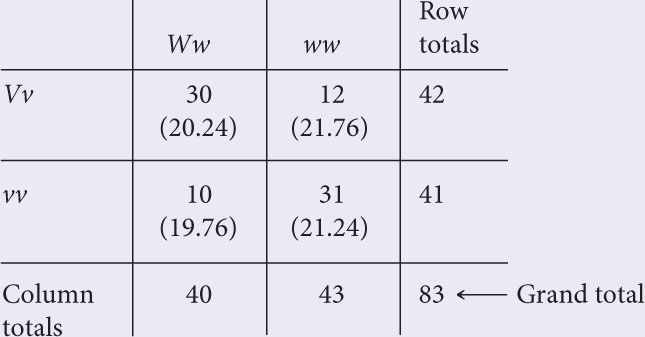
To test the hypothesis, carry out a chi-square test of independence. Construct a table, with the genotypes of the first locus along the top and the genotypes of the second locus along the side. Compute the totals for the rows and columns and the grand total.
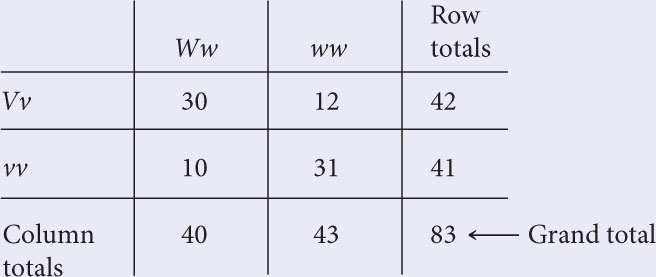
The expected value for each cell of the table is calculated with the following formula:

Using this formula, we find the expected values (given in parentheses) to be:
198
Using these observed and expected numbers, we find the calculated chi-square value to be:

The degrees of freedom for the chi-square test of independence are df = (number of rows − 1) × (number of columns − 1). There are two rows and two columns, and so the degrees of freedom are:
df = (2 − 1) × (2 − 1) = 1 × 1 = 1
In Table 3.5, the probability associated with a chi-square value of 18.39 and 1 degree of freedom is less than 0.005, indicating that chance is very unlikely to be responsible for the differences between the observed numbers and the numbers expected with independent assortment. The genes for coat color and hair type have therefore not assorted independently.
Recall: The probability associated with the chi-square value is the probability that the difference between observed and expected is due to chance.
- b. To determine the recombination frequencies, identify the recombinant progeny. Using the notation for linked genes, write the crosses:The recombination frequency is:
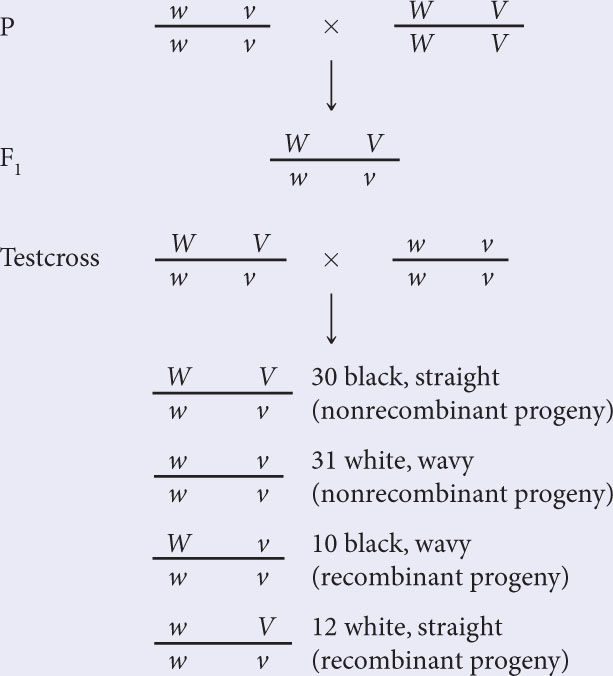
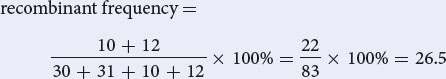
Recall: The recombination frequency =

Problem 2
A series of two-point crosses were carried out among seven loci (a, b, c, d, e, f, and g), producing the following recombination frequencies. Using these recombination frequencies, map the seven loci, showing their linkage groups, the order of the loci in each linkage group, and the distances between the loci of each group:
| Loci | Recombination frequency (%) | Loci | Recombination frequency (%) |
|---|---|---|---|
| a and b | 10 | c and d | 50 |
| a and c | 50 | c and e | 8 |
| a and d | 14 | c and f | 50 |
| a and e | 50 | c and g | 12 |
| a and f | 50 | d and e | 50 |
| a and g | 50 | d and f | 50 |
| b and c | 50 | d and g | 50 |
| b and d | 4 | e and f | 50 |
| b and e | 50 | e and g | 18 |
| b and f | 50 | f and g | 50 |
| b and g | 50 |
Solution Strategy
What information is required in your answer to the problem?
The linkage groups for the seven loci, the order of the loci within each linkage group, and the map distances between the loci.
What information is provided to solve the problem?
Recombination frequencies for each pair of loci.
For help with this problem, review:
Constructing a Genetic Map with the Use of Two-Point Testcrosses in Section 7.2.
199
Solution Steps
To work this problem, remember that 1% recombination equals 1 map unit. The recombination frequency between a and b is 10%; so these two loci are in the same linkage group, approximately 10 m.u. apart.
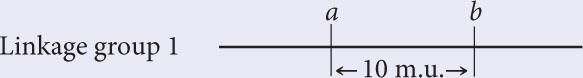
The recombination frequency between a and c is 50%; so c must lie in a second linkage group.
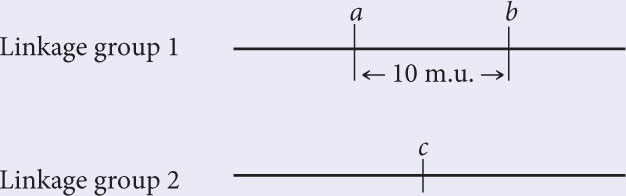
Hint: A recombination frequency of 50% means that genes at the two loci are assorting independently (located in different linkage groups).
The recombination frequency between a and d is 14%; so d is located in linkage group 1. Is locus d 14 m.u. to the right or to the left of locus a? If d is 14 m.u. to the left of a, then the b-to-d distance should be 10 m.u. + 14 m.u. = 24 m.u. On the other hand, if d is to the right of a, then the distance between b and d should be 14 m.u. − 10 m.u. = 4 m.u. The b-d recombination frequency is 4%; so d is 14 m.u. to the right of a. The updated map is:
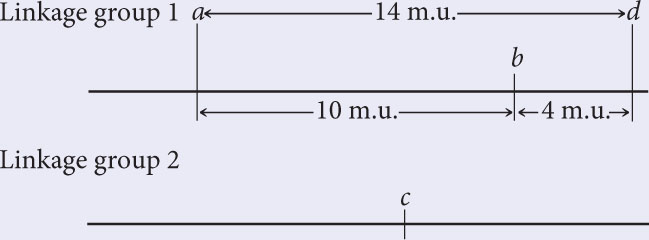
Hint: To determine whether locus d is to the right or the left of locus a, look at the b to d distance.
The recombination frequencies between each of loci a, b, and d, and locus e are all 50%; so e is not in linkage group 1 with a, b, and d. The recombination frequency between e and c is 8 m.u.; so e is in linkage group 2:
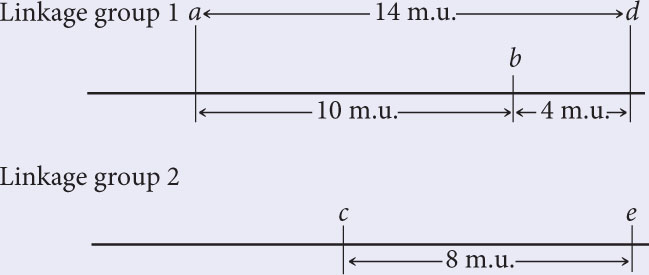
There is 50% recombination between f and all the other genes; so f must belong to a third linkage group:
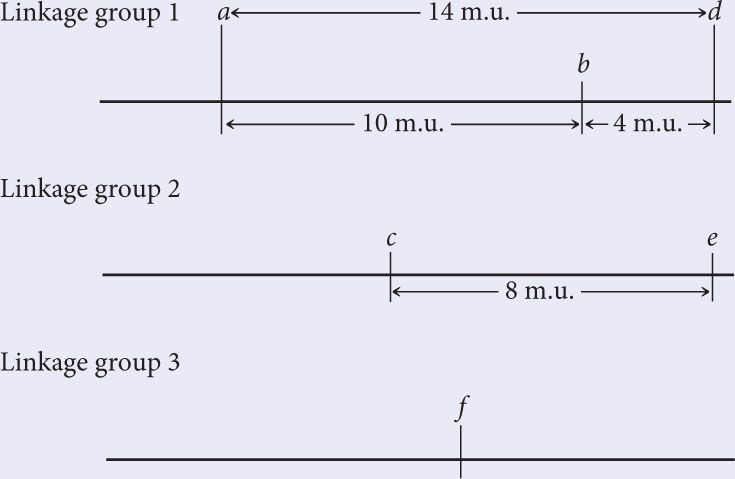
Finally, position locus g with respect to the other genes. The recombination frequencies between g and loci a, b, and d are all 50%; so g is not in linkage group 1. The recombination frequency between g and c is 12 m.u.; so g is a part of linkage group 2. To determine whether g is 12 m.u. to the right or left of c, consult the g–e recombination frequency. Because this recombination frequency is 18%, g must lie to the left of c:
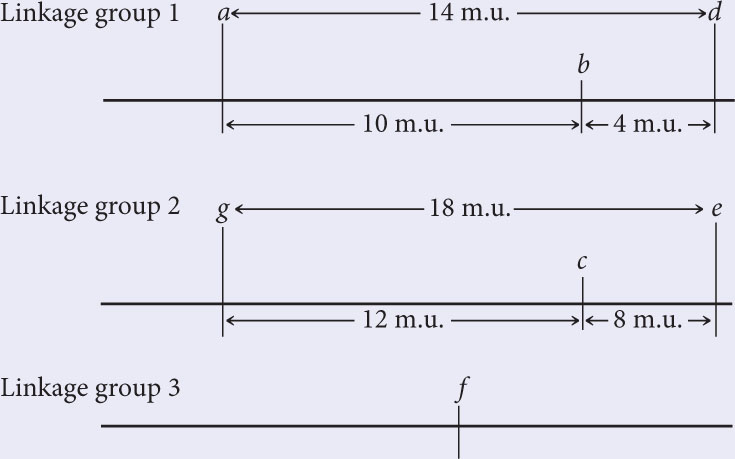
Note that the g-to-e distance (18 m.u.) is shorter than the sum of the g-to-c (12 m.u.) and c-to-e distances (8 m.u.) because of undetectable double crossovers between g and e.
Recall: Because some double crossovers may go undetected, the distance between two distant genes (such as g and e) may be less than the sum of shorter distances (such as g to c and c to e).
200
Problem 3
Ebony body color (e), rough eyes (ro), and brevis bristles (bv) are three recessive mutations that occur in fruit flies. The loci for these mutations have been mapped and are separated by the following map distances:
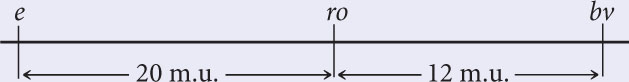
The interference between these genes is 0.4.
A fly with ebony body, rough eyes, and brevis bristles is crossed with a fly that is homozygous for the wild-type traits. The resulting F1 females are test-crossed with males that have ebony body, rough eyes, and brevis bristles; 1800 progeny are produced. Give the expected numbers of phenotypes in the progeny of the testcross.
Solution Strategy
What information is required in your answer to the problem?
The expected numbers of different phenotypes produced by the test cross.
What information is provided to solve the problem?
 The map distances among the three loci.
The map distances among the three loci. The interference among the loci.
The interference among the loci. A testcross is carried out and 1800 progeny are produced
A testcross is carried out and 1800 progeny are produced
For help with this problem, review:
Constructing a Genetic Map with the Three-Point Testcross and the Worked Problem in Section 7.3.
Solution Steps
The crosses are:
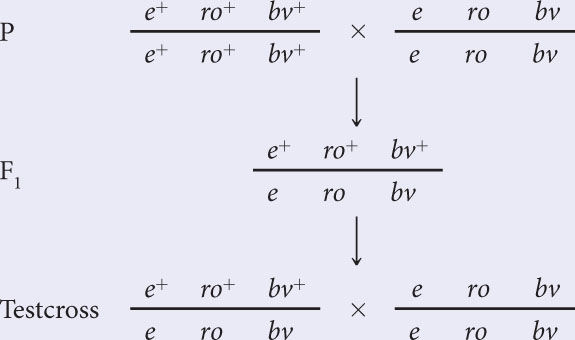
Hint: The order of the genes is provided by the genetic map.
In this case, we know that ro is the middle locus because the genes have been mapped. Eight classes of progeny will be produced from this cross:
| e+ | ro+ | bv+ | nonrecombinant | ||
| e | ro | bv | nonrecombinant | ||
| e+ | / | ro | bv | single crossover between e and ro | |
| e | / | ro+ | bv+ | single crossover between e and ro | |
| e+ | ro+ | / | bv | single crossover between ro and bv | |
| e | ro | / | bv+ | single crossover between ro and bv | |
| e+ | / | ro | / | bv+ | double crossover |
| e | / | ro+ | / | bv | double crossover |
To determine the numbers of each type, use the map distances, starting with the double crossovers. The expected number of double crossovers is equal to the product of the single-crossover probabilities:
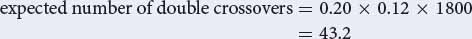
However, there is some interference; so the observed number of double crossovers will be less than the expected. The interference is 1 − coefficient of coincidence; so the coefficient of coincidence is:
coefficient of coincidence = 1 − interference
Recall: The presence of interference means that not all expected double crossovers will be observed.
The interference is given as 0.4; so the coefficient of coincidence equals 1 − 0.4 = 0.6. Recall that the coefficient of coincidence is:
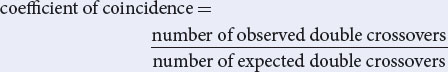
201
Rearranging this equation, we obtain:

number of observed double crossovers = 0.6 × 43.2 = 26
A total of 26 double crossovers should be observed. Because there are two classes of double crossovers ( e+ / ro / bv+ and e / ro+ / bv ), we expect to observe 13 of each class.
Next, we determine the number of single-crossover progeny. The genetic map indicates that the distance between e and ro is 20 m.u.; so 360 progeny (20% of 1800) are expected to have resulted from recombination between these two loci. Some of them will be single-crossover progeny and some will be double-crossover progeny. We have already determined that the number of double-crossover progeny is 26; so the number of progeny resulting from a single crossover between e and ro is 360 − 26 = 334, which will be divided equally between the two single-crossover phenotypes ( e / ro+ / bv+ and e+ / ro bv ).
Hint: To obtain the number of single crossover progeny, subtract the number of double crossover progeny from the total number that resulted from recombination.
The distance between ro and bv is 12 m.u.; so the number of progeny resulting from recombination between these two genes is 0.12 × 1800 = 216. Again, some of these recombinants will be single-crossover progeny and some will be double-crossover progeny. To determine the number of progeny resulting from a single crossover, subtract the double crossovers: 216 − 26 = 190.
Hint: Don’t forget to subtract the double crossovers from the total number of recombinants.
These single-crossover progeny will be divided between the two single-crossover phenotypes ( e+ ro+ / bv an e ro / bv+ ); so there will be 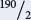 = 95 of each of these phenotypes. The remaining progeny will be nonrecombinants, and they can be obtained by subtraction: 1800 − 26 − 334 − 190 = 1250; there are two nonrecombinants ( e+ ro+ bv+ and e ro bv ); so there will be
= 95 of each of these phenotypes. The remaining progeny will be nonrecombinants, and they can be obtained by subtraction: 1800 − 26 − 334 − 190 = 1250; there are two nonrecombinants ( e+ ro+ bv+ and e ro bv ); so there will be 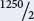 = 625 of each.
= 625 of each.
Hint: The number of nonrecombinants can be obtained by subtraction.
The numbers of the various phenotypes are listed here:
| e+ | ro+ | bv+ | 625 | nonrecombinant | ||
| e | ro | bv | 625 | nonrecombinant | ||
| e+ | / | ro | bv | 167 | single crossover between e and ro | |
| e | / | ro+ | bv+ | 167 | single crossover between e and ro | |
| e+ | ro+ | / | bv | 95 | single crossover between ro and bv | |
| e | ro | / | bv+ | 95 | single crossover between ro and bv | |
| e+ | / | ro | / | bv+ | 13 | double crossover |
| e | / | ro+ | / | bv | 13 | double crossover |
| Total | 1800 | |||||