Chapter 7
Isolate DNA from water samples taken from the two streams. Sequence the DNA and identify the bacterial species based on differences in their DNA.
By using low doses of antibiotics for five years, Farmer Smith selected for bacteria that are resistant to the antibiotics. The doses he used killed sensitive bacteria, but not moderately sensitive or slightly resistant bacteria. As time passed, only resistant bacteria remained in his pigs because any sensitive bacteria were eliminated by the low doses of antibiotics.
In the future, Farmer Smith should continue to use the vitamins, but should use the antibiotics only when a sick pig requires them. In this manner, he will not be selecting for antibiotic-
resistant bacteria, and the chances of successful treatment of his sick pigs will be greater.
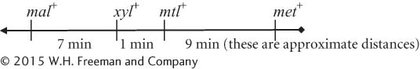
The closer genes are to the F factor, the more quickly they will be transferred. The transfer process will occur in a linear fashion. When conjugation is interrupted, the transfer will stop, and the F− strain will have received only genes carried on the portion of the Hfr strain’s chromosome that entered the F− cell prior to the interruption. From the graph, we can determine when each allele from the Hfr strain was first identified and subsequently approximate the minutes that separate the genes.
The observation that the M and S genes were cotransformed at a high rate suggests that they are located very close to each other on the Streptococcus pneumoniae chromosome.
S and F are cotransformed more frequently (0.22) than are M and F (0.0058), suggesting that S and F are located closer together than are M and F.

Genes M and F cotransform infrequently, which is likely due to the physical distance between them. Genes M and S are more closely linked on chromosome. The relative positions of M, S, and F on the chromosome make it likely that, if M and F are cotransformed, then S will be transformed as well because it is between M and F.
The original infecting phages were wild-
type (a+ b+) and doubly mutant (a− b−). Any phages that give rise to the a+ b− plaque phenotype or the a− b+ plaque phenotype were produced by recombination between the two types of infecting phage particles. Plaque phenotype Number a+ b+ 2043 a+ b− 320 (recombinant) a− b+ 357 (recombinant) a− b− 2134 Total plaques 4854 The frequency of recombination is calculated by dividing the total number of recombinant plaques by the total number of plaques (677/4854), which gives a frequency of 0.14, or 14%.
Transductants were initially screened for the presence of proC+. Thus, only proC+ transductants were identified.
The wild-
type genotypes (proC+ proA+ proB+ proD+) represent single transductants of proC+. Both the proC+ proA− proB+ proD+ and proC+ proA+ proB– proD+ genotypes represent cotransductants of proC+, proA− and proC+, proB−. Both proA and proB were cotransduced with proC at about the same rate, which suggests that they are similar in distance to proC. The proD gene was never cotransduced with proC, suggesting that proD is more distant from proC than proA and proB. However, there were a small number of all types of cotransductants, and the absence of cotransduction between proD and proC might be due to chance.
Number of plaques produced by c+ m+ = 460; by c− m− = 460; by c+ m− = 40 (recombinant); by c− m+ = 40 (recombinant).
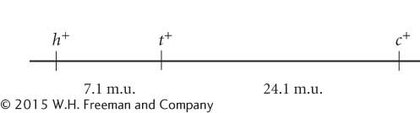
Coefficient of coincidence = (6 + 5)/(0.071 × 0.241 × 942) = 0.68. Interference = 1 − 0.68 = 0.32.