Messenger RNA Processing
Messenger RNA functions as the template for protein synthesis; it carries genetic information from DNA to a ribosome and helps to assemble amino acids in their correct order. In bacteria, mRNA is transcribed directly from DNA, but in eukaryotes, a pre-mRNA (the primary transcript) is first transcribed from DNA and then processed to yield the mature mRNA. We will reserve the term “mRNA” for RNA molecules that have been completely processed and are ready to undergo translation.
THE STRUCTURE OF mRNA In mRNA, each amino acid in a protein is specified by a set of three nucleotides called a codon. Both prokaryotic and eukaryotic mRNAs contain three primary regions (Figure 10.17). The 5′ untranslated region (5′ UTR; sometimes called the leader), a sequence of nucleotides at the 5′ end of the mRNA, does not encode any of the amino acids of a protein. In bacterial mRNA, this region contains a consensus sequence (UAAGGAGGU) called the Shine–Dalgarno sequence, which serves as the ribosome-binding site during translation; it is found approximately seven nucleotides upstream of the first codon translated into an amino acid (called the start codon). The Shine–Dalgarno sequence is complementary to sequences found in one of the RNA molecules that make up the ribosome and pairs with those sequences during translation. In eukaryotic cells, ribosomes bind to a modified 5′ end of mRNA, as discussed later in this section.
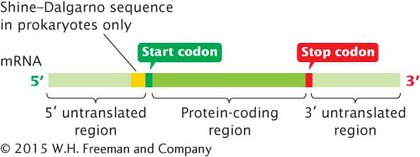
Figure 10.17: The three primary regions of mature mRNA are the 5′ untranslated region, the protein-coding region, and the 3′ untranslated region.
The next section of mRNA is the protein-coding region, which comprises the codons that specify the amino acid sequence of the protein. The protein-coding region begins with a start codon and ends with a stop codon.
The last region of mRNA is the 3′ untranslated region (3′ UTR; sometimes called a trailer), a sequence of nucleotides that is at the 3′ end of the mRNA and is not translated into protein. The 3′ untranslated region affects the stability of mRNA and the translation of the mRNA protein-coding sequence. View Animation 10.2 to see how mutations in different regions of a gene affect the flow of information from genotype to phenotype.
CONCEPTS
Messenger RNA molecules contain three main regions: a 5′ untranslated region, a protein-coding region, and a 3′ untranslated region. The 5′ and 3′ untranslated regions do not encode any amino acids of a protein, but contain information that is important in translation and RNA stability.
In bacterial cells, transcription and translation take place simultaneously: while the 3′ end of an mRNA is undergoing transcription, ribosomes attach to the Shine—Dalgarno sequence near the 5′ end and begin translation. Because transcription and translation are coupled, there is little opportunity for the bacterial mRNA to be modified before protein synthesis. In contrast, transcription and translation are separated in both time and space in eukaryotic cells. Transcription takes place in the nucleus, whereas translation takes place in the cytoplasm; this separation provides an opportunity for eukaryotic RNA to be modified before it is translated. Indeed, eukaryotic mRNA is extensively altered after transcription. Changes are made to the 5′ end, the 3′ end, and the protein-coding section of the RNA molecule.
ADDITION OF THE 5′ CAP One type of modification of eukaryotic pre-mRNA is the addition of a structure called a 5′ cap. The cap consists of an extra modified nucleotide at the 5′ end of the mRNA as well as methyl groups (CH3) on the 2′-OH group of the sugar of one or more nucleotides at the 5′ end (Figure 10.18). The addition of the cap takes place rapidly after the initiation of transcription. The cap functions in the initiation of translation, as we’ll see in Chapter 11. Cap-binding proteins recognize the cap and attach to it; a ribosome then binds to these proteins and moves downstream along the mRNA until the start codon is reached and translation begins. The presence of a 5′ cap also increases the stability of mRNA and influences the removal of introns.
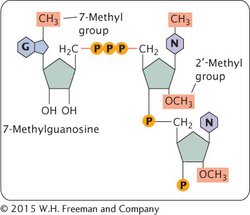
Figure 10.18: Most eukaryotic mRNAs have a 5′ cap. The cap consists of a nucleotide with 7-methylguanine that is attached to the 5′ end of pre-mRNA by a unique 5′—5′ bond, as well as methyl groups added to the 2′ position of the sugars in the second and third nucleotides of the pre-mRNA, and sometimes a methyl group added to the base (N) on the initial nucleotide.
ADDITION OF THE POLY(A) TAIL A second type of modification to eukaryotic mRNA is the addition of 50–250 or more adenine nucleotides at the 3′ end, forming a poly(A) tail. These nucleotides are not encoded in the DNA, but are added after transcription (Figure 10.19) in a process termed polyadenylation. Many eukaryotic genes are transcribed well beyond the end of the coding sequence; most of the extra material at the 3′ end is then cleaved, and the poly(A) tail is added. For some pre-mRNA molecules, more than 1000 nucleotides may be removed from the 3′ end before polyadenylation.
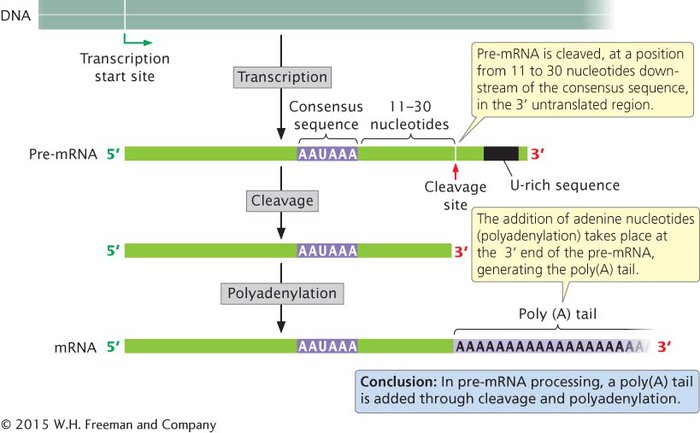
Figure 10.19: Most eukaryotic mRNAs have a 3′ poly(A) tail.
Processing of the 3′ end of pre-mRNA requires sequences, termed the polyadenylation signal, located upstream and downstream of the site where cleavage occurs. The consensus sequence AAUAAA is usually from 11 to 30 nucleotides upstream of the cleavage site (see Figure 10.19) and determines the point at which cleavage will take place. A sequence rich in uracil nucleotides (or guanine and uracil nucleotides) is typically downstream of the cleavage site. A large number of proteins take part in finding the cleavage site and removing the 3′ end. After cleavage has been completed, adenine nucleotides are added without a template to the new 3′ end, creating the poly(A) tail.
The poly(A) tail confers stability on many mRNAs, increasing the time during which the mRNA remains intact and available for translation before it is degraded by cellular enzymes. The stability conferred by the poly(A) tail depends on the proteins that attach to the tail and on its length. The poly(A) tail also facilitates the attachment of the ribosome to the mRNA and plays a role in export of the mRNA into the cytoplasm.
CONCEPTS
Eukaryotic pre-mRNAs are processed at their 5′ and 3′ ends. A 5′ cap, consisting of a modified nucleotide and several methyl groups, is added to the 5′ end. The cap facilitates the binding of a ribosome, increases the stability of the mRNA, and may affect the removal of introns. Processing at the 3′ end includes cleavage downstream of an AAUAAA consensus sequence and the addition of a poly(A) tail.
RNA SPLICING The other major type of modification of eukaryotic pre-mRNA is the removal of introns by RNA splicing. This modification takes place in the nucleus, before the RNA moves to the cytoplasm. Splicing requires the presence of three sequences in the intron. One end of the intron is referred to as the 5′ splice site, and the other end is the 3′ splice site (Figure 10.20); these splice sites possess short consensus sequences. Most introns in pre-mRNAs begin with GU and end with AG, indicating that these sequences play a crucial role in splicing. Indeed, changing a single nucleotide at either of these sites prevents splicing.
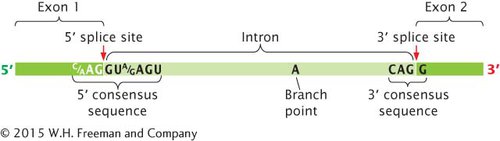
Figure 10.20: The splicing of pre-mRNA requires consensus sequences. Critical consensus sequences are present at the 5′ splice site and the 3′ splice site. A weak consensus sequence (not shown) exists at the branch point.
The third sequence important for splicing is at the branch point, which is an adenine nucleotide that lies from 18 to 40 nucleotides upstream of the 3′ splice site (see Figure 10.20). The sequence surrounding the branch point is a weak consensus sequence. Deletion or mutation of the adenine nucleotide at the branch point prevents splicing.
Splicing takes place within a large structure called the spliceosome, which is one of the largest and most complex of all molecular structures. The spliceosome consists of five RNA molecules and almost three hundred proteins. The RNA components are small nuclear RNAs; these snRNAs associate with proteins to form small nuclear ribonucleoprotein particles (snRNPs). Each snRNP contains a single snRNA molecule and multiple proteins. The spliceosome is composed of five snRNPs (U1, U2, U4, U5, and U6) and some proteins not associated with an snRNA.
CONCEPTS
Introns in pre-mRNAs contain three consensus sequences critical to splicing: a 5′ splice site, a 3′ splice site, and a branch point. The splicing of pre-mRNA takes place within a large complex called the spliceosome, which consists of snRNAs and proteins.
 CONCEPT CHECK 5
CONCEPT CHECK 5
If a splice site were mutated so that splicing did not take place, what would the effect be on the mRNA?
It would be shorter than normal.
It would be longer than normal.
It would be the same length but would encode a different protein.
Before splicing takes place, an intron lies between an upstream exon (exon 1) and a downstream exon (exon 2), as shown in Figure 10.21. Pre-mRNA is spliced in two distinct steps. In the first step of splicing, the pre-mRNA is cut at the 5′ splice site. This cut frees exon 1 from the intron, and the 5′ end of the intron attaches to the branch point; that is, the intron folds back on itself, forming a structure called a lariat. In this reaction, the guanine nucleotide in the consensus sequence at the 5′ splice site bonds with the adenine nucleotide at the branch point through a transesterification reaction. As a result, the 5′ phosphate group of the guanine nucleotide becomes attached to the 2′-OH group of the adenine nucleotide at the branch point (see Figure 10.21).
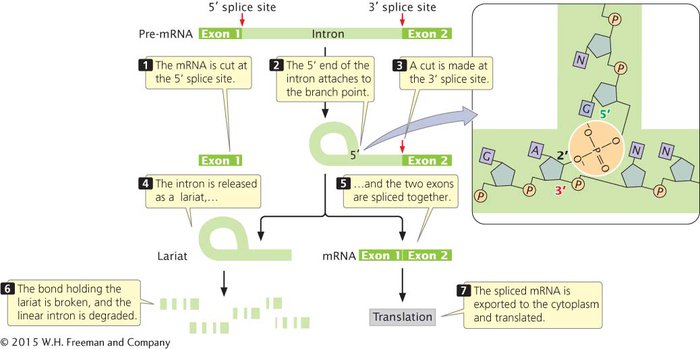
Figure 10.21: The splicing of pre-mRNA introns requires a two-step process.
In the second step of RNA splicing, a cut is made at the 3′ splice site, and simultaneously, the 3′ end of exon 1 becomes covalently attached (spliced) to the 5′ end of exon 2. The intron is released as a lariat. Eventually, a lariat debranching enzyme breaks the bond at the branch point, producing a linear intron that is rapidly degraded by nuclear enzymes. The mature mRNA, consisting of the exons spliced together, is exported to the cytoplasm, where it is translated. These splicing reactions take place within the spliceosome, which carries out the splicing reactions.
Many eukaryotic mRNAs undergo alternative processing, in which a single pre-mRNA is processed in different ways to produce alternative types of mRNA, resulting in the production of different proteins from the same DNA sequence. One type of alternative processing is alternative splicing, in which the same pre-mRNA can be spliced in more than one way to yield multiple mRNAs that are translated into different amino acid sequences and thus different proteins (Figure 10.22). Alternative processing is an important source of protein diversity in vertebrates; an estimated 60% of all human genes are alternatively spliced.
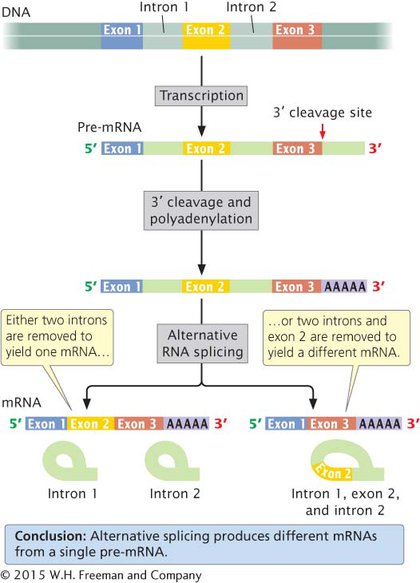
Figure 10.22: Eukaryotic cells have alternative pathways for processing pre-mRNA.
CONCEPTS
Intron splicing in pre-mRNAs is a two-step process: (1) the 5′ end of an intron is cleaved and attached to the branch point to form a lariat, and (2) the 3′ end of the intron is cleaved and the two ends of the exon are spliced together. These reactions take place within the spliceosome. Alternative splicing enables exons to be spliced together in different combinations to yield mRNAs that encode different proteins.
CONNECTING CONCEPTS
Eukaryotic Gene Structure and Pre-mRNA Processing
This chapter has introduced a number of different components of genes and RNA molecules, including promoters, 5′ untranslated regions, coding sequences, exons, introns, 3′ untranslated regions, poly(A) tails, and caps. Let’s see how some of these components are combined to create a typical eukaryotic gene and how a mature mRNA is produced from them.
The promoter, which typically lies upstream of the transcription start site, is necessary for transcription to take place, but is itself not usually transcribed when protein-encoding genes are transcribed by RNA polymerase II (Figure 10.23a). Farther upstream or downstream of the start site there may be sequences called enhancers, DNA sequences that also regulate transcription. In transcription, all the nucleotides between the transcription start site and the termination site are transcribed into pre-mRNA, including exons, introns, and a long 3′ end that is later cleaved from the transcript (Figure 10.23b). Notice that the 5′ end of the first exon contains the sequence that encodes the 5′ untranslated region, and that the 3′ end of the last exon contains the sequence that encodes the 3′ untranslated region.
The pre-mRNA is then processed to yield a mature mRNA. The first step in this processing is the addition of a cap to the 5′ end of the pre-mRNA (Figure 10.23c). Next, the 3′ end is cleaved at a site downstream of the AAUAAA consensus sequence in the last exon (Figure 10.23d). Immediately after cleavage, a poly(A) tail is added to the 3′ end (Figure 10.23e). Finally, the introns are removed to yield the mature mRNA (Figure 10.23f). The mRNA now contains 5′ and 3′ untranslated regions, which are not translated into amino acids, and the nucleotides that carry the protein-coding sequences. You can explore the consequences of failed RNA processing by viewing and interacting with Animation 10.3.
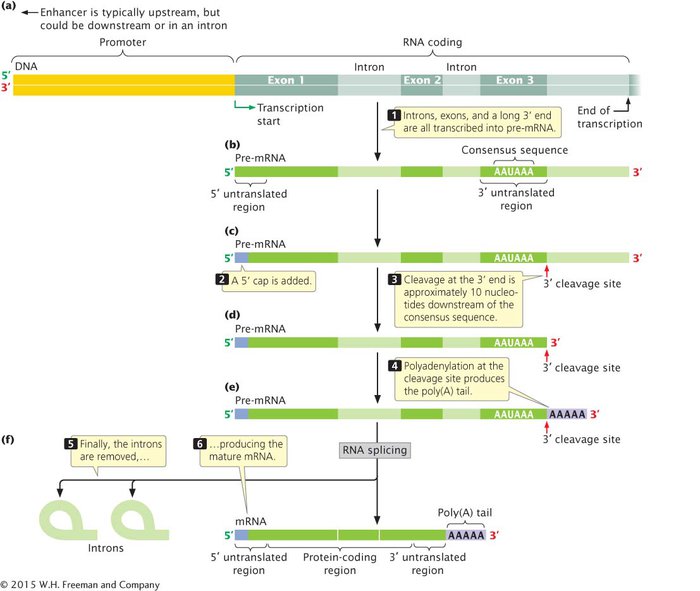
Figure 10.23: Mature eukaryotic mRNA is produced when pre-mRNA is transcribed and undergoes several types of processing.




 CONCEPT CHECK 5
CONCEPT CHECK 5

