Small RNA Molecules and RNA Interference
Numerous small RNA molecules (most of them 20–30 nucleotides long) have been discovered that greatly influence many basic biological processes, including the formation of chromatin structure, transcription, and translation. These small RNA molecules play important roles in gene expression, development, cancer, and defense against foreign DNA. They are also being harnessed by researchers to study gene function and treat genetic diseases. The discovery of small RNA molecules has greatly influenced our understanding of how genes are regulated and has demonstrated the importance of DNA sequences that do not encode proteins.
In 1998, Andrew Fire, Craig Mello, and their colleagues observed a strange phenomenon. They were inhibiting the expression of genes in the nematode Caenorhabditis elegans by injecting single-stranded RNA molecules that were complementary to a gene’s DNA sequence. Called antisense RNA, such molecules are known to inhibit gene expression by binding to mRNA sequences and inhibiting translation. However, Fire, Mello, and their colleagues found that even more potent gene silencing was triggered when double-stranded RNA was injected into the animals. This finding was puzzling because no mechanism by which double-stranded RNA could inhibit translation was known. Several other, previously described types of gene silencing were also found to be triggered by double-stranded RNA.
Subsequent research revealed an astonishing array of small RNA molecules with important cellular functions in eukaryotes, which now include at least three major classes: small interfering RNAs (siRNAs), microRNAs (miRNAs), and Piwi-interacting RNAs (piRNAs). These small RNAs are found in many eukaryotes and are responsible for a variety of different functions (see Table 10.2). An analogous group of small RNAs with silencing functions—CRISPR RNAs (crRNAs)—have been detected in prokaryotes. For their discovery of RNA interference, Fire and Mello were awarded the Nobel Prize in Physiology or Medicine in 2006.
RNA interference (RNAi) is a powerful and precise mechanism used by eukaryotic cells to limit the invasion of foreign genes (from viruses and transposons) and to censor the expression of their own genes. RNA interference is triggered by double-stranded RNA molecules, which may arise in several ways (Figure 10.25): by the transcription of inverted repeats into an RNA molecule that then base pairs with itself to form double-stranded RNA; by the simultaneous transcription of two different RNA molecules that are complementary to one another and that pair, forming double-stranded RNA; or by infection by viruses that make double-stranded RNA. These double-stranded RNA molecules are chopped up by an enzyme appropriately called Dicer, resulting in tiny RNA molecules that are unwound to produce siRNAs and miRNAs (see Figure 10.25).
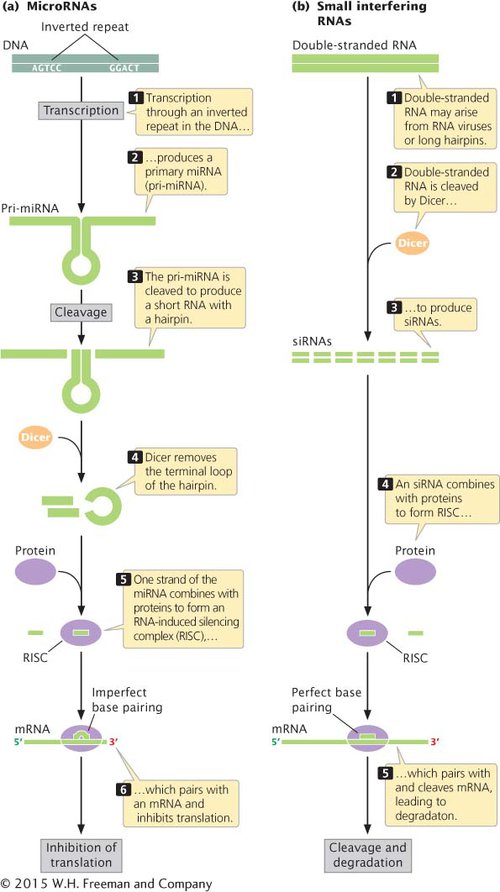
Figure 10.25: Small interfering RNAs and microRNAs are produced from double-stranded RNAs.
RNA interference is responsible for regulating a number of key genetic and developmental processes, including changes in chromatin structure, translation, cell fate and proliferation, and cell death. Geneticists also use the RNAi machinery as an effective tool for blocking the expression of specific genes (see Chapter 14).
SMALL INTERFERING AND microRNAs Two abundant classes of RNA molecules that function in RNA interference in eukaryotes are small interfering RNAs and microRNAs. Although these two types of RNA differ in how they originate (Table 10.5; see also Figure 10.25), they have a number of features in common, and their functions overlap considerably.
TABLE 10.5 Differences between siRNAs and miRNAs
| Feature |
siRNA |
miRNA |
| Origin |
mRNA, transposon, or virus |
RNA transcribed from distinct gene |
| Cleavage of |
RNA duplex or single-stranded RNA that forms long hairpins |
Single-stranded RNA that forms short hairpins of double-stranded RNA |
| Action |
Degradation of mRNA, inhibition of transcription, chromatin modification |
Degradation of mRNA, inhibition of translation, chromatin modification |
| Target |
Genes from which they were transcribed |
Genes other than those from which they were transcribed |
Both siRNA and miRNA molecules combine with proteins to form an RNA-induced silencing complex (RISC) (see Figure 10.25). Key to the functioning of RISCs is a protein called Argonaute. The RISC pairs with an mRNA molecule that possesses a sequence complementary to the RISC’s siRNA or miRNA component, and then either cleaves the mRNA, leading to degradation of the mRNA, or represses translation of the mRNA. Some siRNAs also serve as guides for the methylation of complementary sequences in DNA, whereas others alter chromatin structure, both of which affect transcription.
MicroRNAs have been found in all eukaryotic organisms examined to date as well as in viruses: they control the expression of genes taking part in many biological processes, including growth, development, and metabolism. To see how small interfering RNAs and microRNAs affect gene expression, see Animation 10.4.
THE PROCESSING OF microRNAS AND siRNAs The genes that encode miRNA are transcribed into longer precursors, called primary microRNA (pri-miRNA), that range from several hundred to several thousand nucleotides in length (see Figure 10.25a). The pri-miRNA is then cleaved into one or more smaller RNA molecules with a hairpin, which is a secondary structure that forms when sequences on the same strand are complementary and pair with one another. Dicer binds to this hairpin structure and removes the terminal loop. One of the miRNA strands is incorporated into the RISC; the other strand is released and degraded. The RISC then attaches to a complementary sequence on the target mRNA, usually in the 3′ untranslated region of the mRNA.
Small interfering RNAs are processed in a similar way (see Figure 10.25b). Double-stranded RNA from viruses or long hairpins is cleaved by Dicer to produce siRNAs, which combine with proteins to form RISC. The RISC then pairs with sequences on the target mRNA and cleaves the mRNA, after which the mRNA is degraded.
CONCEPTS
Small interfering RNAs and microRNAs are tiny RNAs produced when larger double-stranded RNA molecules are cleaved by the enzyme Dicer. Small interfering RNAs and microRNAs participate in a variety of processes, including mRNA degradation, the inhibition of translation, the methylation of DNA, and chromatin remodeling.
