Breaking the Genetic Code
In 1953, James Watson and Francis Crick solved the structure of DNA and identified its base sequence as the carrier of genetic information (as we saw in Chapter 8). However, the way in which the base sequence of DNA specifies the amino acid sequences of proteins (the genetic code) remained elusive for another 10 years.
One of the first questions about the genetic code to be addressed was how many nucleotides are necessary to specify a single amino acid. The set of nucleotides that encodes a single amino acid— TRY PROBLEMS 12 AND 13
TRY PROBLEMS 12 AND 13
CONCEPTS
The genetic code is a triplet code, in which three nucleotides encode each amino acid in a protein.
 CONCEPT CHECK 2
CONCEPT CHECK 2
A codon is
one of three nucleotides that encode an amino acid.
three nucleotides that encode an amino acid.
three amino acids that encode a nucleotide.
one of four bases in DNA.
b
Once it had been firmly established that the genetic code consists of codons that are three nucleotides in length, the next step was to determine which groups of three nucleotides specify which amino acids. Logically, the easiest way to break the code would have been to determine the base sequence of a piece of RNA, add it to a test tube containing all the components necessary for translation, and allow it to direct the synthesis of a protein. The amino acid sequence of the newly synthesized protein could then be determined, and its sequence could be compared with that of the RNA. Unfortunately, there was no way at that time to determine the nucleotide sequence of a piece of RNA, so indirect methods were necessary to break the code.
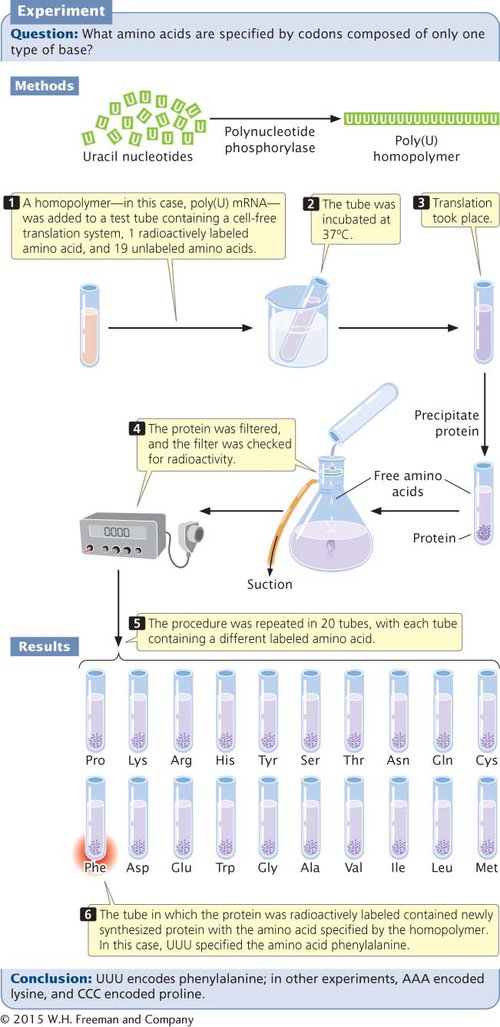
The first clues to the genetic code came in 1961, from the work of Marshall Nirenberg and Johann Heinrich Matthaei. These investigators created synthetic RNAs by using an enzyme called polynucleotide phosphorylase. Unlike RNA polymerase, polynucleotide phosphorylase does not require a template; it randomly links together any RNA nucleotides that happen to be available. The first synthetic mRNAs used by Nirenberg and Matthaei were homopolymers, RNA molecules consisting of a single type of nucleotide. For example, by adding polynucleotide phosphorylase to a solution of uracil nucleotides, they generated RNA molecules that consisted entirely of uracil nucleotides and thus contained only UUU codons (Figure 11.4). These poly(U) RNAs were then added to 20 test tubes, each containing the components necessary for translation and all 20 amino acids. A different amino acid was radioactively labeled in each of the 20 tubes. Radioactive protein appeared in only one of the tubes: the one containing labeled phenylalanine (see Figure 11.4). This result showed that the codon UUU specifies the amino acid phenylalanine. The results of similar experiments using poly(C) and poly(A) RNA demonstrated that CCC encodes proline and AAA encodes lysine; for technical reasons, the results from poly(G) were uninterpretable. Other experiments provided additional information about the genetic code, and it was fully understood by 1968.
293