RNA Interference and Gene Regulation
The expression of a number of eukaryotic genes is controlled through RNA interference, also known as RNA silencing and posttranscriptional gene silencing (see Chapter 10). Research suggests that as many as 30% of human genes are regulated by RNA interference. RNA interference is widespread in eukaryotes, existing in fungi, plants, and animals. A similar group of small RNA molecules with silencing and activating functions have been detected in prokaryotes. The mechanism of RNA silencing is also widely used as a powerful technique for artificially regulating gene expression in genetically engineered organisms (see Chapter 14).
329
RNA interference is triggered by small RNA molecules know as microRNAs (miRNAs) and small interfering RNAs (siRNAs), depending on their origin and mode of action (see Chapter 10). An enzyme called Dicer cleaves and processes double-
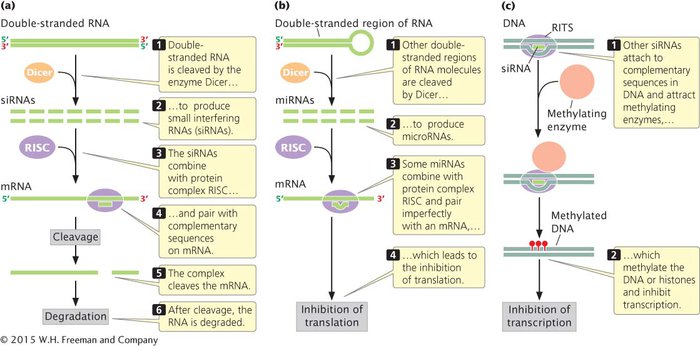
RNA CLEAVAGE RISCs that contain an siRNA (and some that contain an miRNA) pair with mRNA molecules and cleave the mRNA near the middle of the bound siRNA (Figure 12.21a). This cleavage is carried out by a protein that is sometimes referred to as Slicer. After cleavage, the mRNA is further degraded. Thus, the presence of siRNAs and miRNAs increases the rate at which mRNAs are broken down and decreases the amount of protein produced.
INHIBITION OF TRANSLATION Some miRNAs regulate genes by inhibiting the translation of their complementary mRNAs (Figure 12.21b). For example, an important gene in flower development in Arabidopsis thaliana is APETALA2. The expression of this gene is regulated by an miRNA that base pairs with nucleotides in the coding region of APETALA2 mRNA and inhibits its translation.
TRANSCRIPTIONAL SILENCING Other siRNAs silence transcription by altering chromatin structure. These siRNAs combine with proteins to form a complex called RITS (for RNA-
CONCEPTS
RNA interference is initiated by double-
 CONCEPT CHECK 10
CONCEPT CHECK 10
In RNA silencing, siRNAs and miRNAs usually bind to which part of the mRNA molecules that they control?
5′ UTR
5′ cap
3′ poly(A) tail
3′ UTR
d