APPLICATION QUESTIONS AND PROBLEMS
Introduction
Question 14
14.The introduction to this chapter discusses the use of gene therapy to treat Leber congenital amaurosis (LCA, a type of severe blindness). What characteristics of blindness that affects the retina (such as LCA) make it an attractive candidate for treatment by gene therapy?
Section 14.1
Question 15
*15.Suppose that a geneticist discovers a new restriction enzyme in the bacterium Aeromonas ranidae. This restriction enzyme is the first to be isolated from this bacterial species. Using the standard convention for abbreviating restriction enzymes, give this new restriction enzyme a name (for help, see the footnote to Table 14.1).
Question 16
16.How often, on average, would you expect a restriction endonuclease to cut a DNA molecule if the recognition sequence for the enzyme had 5 bp? (Assume that the four types of bases are equally likely to be found in the DNA and that the bases in a recognition sequence are independent.) How often would the endonuclease cut the DNA if the recognition sequence had 8 bp?
Question 17
*17.A microbiologist discovers a new restriction endonuclease. When DNA is digested by this enzyme, fragments that average 1,048,500 bp in length are produced. What is the most likely number of base pairs in the recognition sequence of this enzyme?
Question 18
18.Will restriction sites for an enzyme that has 4 bp in its recognition sequence be closer together, farther apart, or similarly spaced, on average, compared with those of an enzyme that has 6 bp in its recognition sequence? Explain your reasoning.
Question 19
*19.About 60% of the base pairs in a human DNA molecule are AT. If the human genome has 3.2 billion base pairs of DNA, about how many times will the following restriction sites be present?
BamHI (recognition sequence is 5′—GGATCC—
3′) EcoRI (recognition sequence is 5′—GAATTC—
3′) HaeIII (recognition sequence is 5′—GGCC—
3′)
Question 20
*20.A linear piece of DNA has the following EcoRI restriction sites:
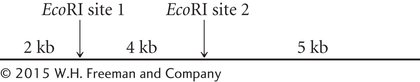
This piece of DNA is cut by EcoRI, the resulting fragments are separated by gel electrophoresis, and the gel is stained with ethidium bromide. Draw a picture of the bands that will appear on the gel.
400
If a mutation that alters EcoRI site 1 occurs in this piece of DNA, how will the banding pattern on the gel differ from the one that you drew in part a?
If mutations that alter EcoRI sites 1 and 2 occur in this piece of DNA, how will the banding pattern on the gel differ from the one that you drew in part a?
If 1000 bp of DNA were inserted between the two restriction sites, how would the banding pattern on the gel differ from the one that you drew in part a?
If 500 bp of DNA between the two restriction sites were deleted, how would the banding pattern on the gel differ from the one that you drew in part a?
Question 21
*21.Which vectors (plasmid, phage λ, cosmid, bacterial artificial chromosome) can be used to clone a continuous fragment of DNA with the following lengths?
4 kb
20 kb
35 kb
100 kb
Question 22
22.A geneticist uses a plasmid for cloning that has the lacZ gene and a gene that confers resistance to penicillin. The geneticist inserts a piece of foreign DNA into a restriction site that is located within the lacZ gene and uses the plasmid to transform bacteria. Explain how the geneticist can identify bacteria that contain a copy of a plasmid with the foreign DNA.
Section 14.2
Question 23
23.Suppose that you have just graduated from college and have started working at a biotechnology firm. Your first assignment is to clone the pig gene for the hormone prolactin. Assume that the pig gene for prolactin has not yet been isolated, sequenced, or mapped; however, the mouse gene for prolactin has been cloned and the amino acid sequence of mouse prolactin is known. Briefly explain two different strategies that you might use to find and clone the pig gene for prolactin.
Section 14.3
Question 24
24.In Figure 14.11, Bob and Joe are each homozygous for different restriction fragment patterns. How many bands would you expect to see on the gel if a person was heterozygous for the A and B patterns? Explain your reasoning.
Question 25
25.Suppose that you want to sequence the following DNA fragment:
5′—TCCCGGGAAA-
You first use PCR to amplify the fragment, so that there is sufficient DNA for sequencing. You carry out dideoxy sequencing of the fragment. You then separate the products of the polymerization reactions by gel electrophoresis. Draw the bands that should appear on the gel from the four sequencing reactions.
Question 26
*26.Suppose that you are given a short fragment of DNA to sequence. You amplify the fragment with PCR and set up a series of four dideoxy reactions. You then separate the products of the reactions by gel electrophoresis and obtain the following banding pattern:
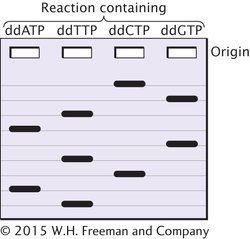
Write out the base sequence of the original fragment that you were given.
Original sequence: 5′— __________________ —3′
Question 27
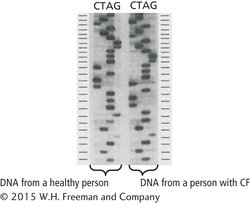
27. The picture below is a sequencing gel from the original study that first sequenced the cystic fibrosis gene (J. R. Riordan et al. 1989. Science 245:1066–1073). From the picture, determine the sequence of the normal copy of the gene and the sequence of the mutated copy of the gene. Identify the location of the mutation that causes cystic fibrosis (CF). Hint: The CF mutation is a 3 bp deletion.
The picture below is a sequencing gel from the original study that first sequenced the cystic fibrosis gene (J. R. Riordan et al. 1989. Science 245:1066–1073). From the picture, determine the sequence of the normal copy of the gene and the sequence of the mutated copy of the gene. Identify the location of the mutation that causes cystic fibrosis (CF). Hint: The CF mutation is a 3 bp deletion.
Section 14.4
Question 28
28.You have discovered a gene in mice that is similar to a gene in yeast. How might you determine whether this gene is essential for development in mice?