Cutting and Joining DNA Fragments
A first step in the molecular analysis of a DNA segment or gene is to isolate it from the remainder of the DNA. A key moment in the development of molecular genetic methods was the discovery in the late 1960s of restriction enzymes (also called restriction endonucleases), which recognize and make double-
More than 800 different restriction enzymes that recognize and cut DNA at more than 100 different sequences have been isolated from bacteria. Many of these enzymes are commercially available; examples of some commonly used restriction enzymes are given in Table 14.1. The name of each restriction enzyme begins with an abbreviation that signifies its bacterial origin.
| Enzyme | Microorganism from which enzyme is produced |
Recognition sequence | Type of fragment end produced |
|---|---|---|---|
| BamHI | Bacillus amyloliquefaciens |  |
Cohesive |
| CofI | Clostridium formicoaceticum | 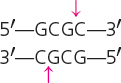 |
Cohesive |
| EcoRI | Escherichia coli | 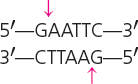 |
Cohesive |
| EcoRII | Escherichia coli | 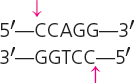 |
Cohesive |
| HaeIII | Haemophilus aegyptius | 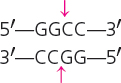 |
Blunt |
| HindIII | Haemophilus influenzae | 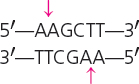 |
Cohesive |
| PvuII | Proteus vulgaris |  |
Blunt |
|
Note: The first three letters of the abbreviation for each restriction enzyme refer to the bacterial species from which the enzyme was isolated (e.g., Eco refers to E. coli). A fourth letter may refer to the strain of bacteria from which the enzyme was isolated (the “R” in EcoRI indicates that this enzyme was isolated from the RY13 strain of E. coli). Roman numerals that follow the letters identify different enzymes from the same species. |
|||
374
The sequences recognized by restriction enzymes are usually from 4 to 8 bp long; most enzymes recognize a sequence of 4 or 6 bp. Most recognition sequences are palindromic—
Some restriction enzymes make staggered cuts in the DNA. For example, HindIII recognizes the following sequence:
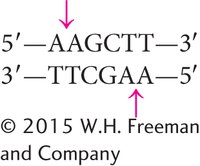
HindIII cuts the sugar–phosphate backbone of each strand at the point indicated by the arrow, generating fragments with short, single-

Such ends are called cohesive ends, or sticky ends, because they are complementary to each other and can spontaneously pair to connect the fragments. Thus, DNA fragments with sticky ends can be “glued” together: any two fragments cleaved by the same enzyme will have complementary ends and will pair (Figure 14.1). When their cohesive ends have paired, the DNA fragments can be joined together permanently by DNA ligase, which seals nicks between the sugar–phosphate groups of the fragments.
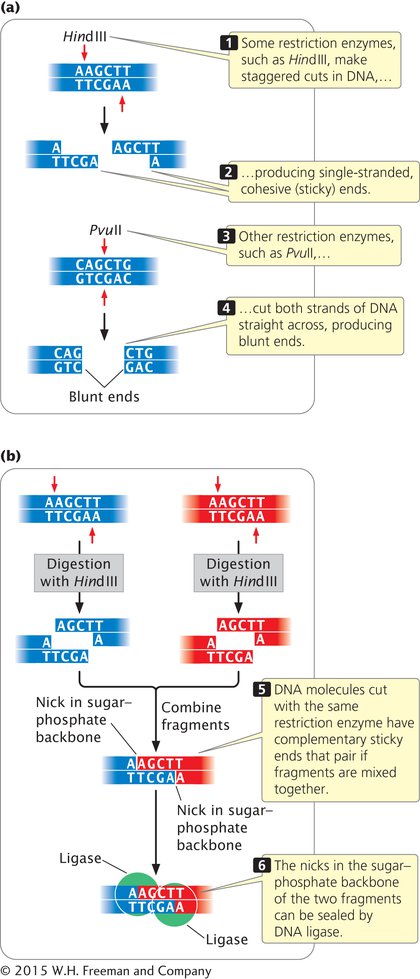
Not all restriction enzymes produce staggered cuts and sticky ends (see Figure 14.1a). PvuII cuts in the middle of its recognition sequence, and the cuts on the two strands are directly opposite one another, producing blunt-
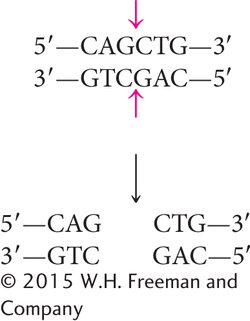
Fragments that have blunt ends are usually joined together in other ways.
The sequences recognized by a restriction enzyme are located randomly within the genome. Consequently, there is a relation between the length of the recognition sequence and the number of times it is present in a genome: there will be fewer longer recognition sequences than shorter recognition sequences because the probability of the occurrence of a particular sequence consisting of, say, six specific bases is less than the probability of the occurrence of a particular sequence of four specific bases. Therefore, restriction enzymes that recognize longer sequences will cut a given piece of DNA into fewer and longer fragments than will restriction enzymes that recognize shorter sequences.
Restriction enzymes are used whenever DNA fragments must be cut or joined. In a typical restriction reaction, a concentrated solution of purified DNA is placed in a small tube with a buffer solution and a small amount of restriction enzyme. Within a few hours, the enzyme cuts the appropriate restriction sites in all the DNA molecules, producing a mixture of DNA fragments.
375
In recent years, geneticists have designed more complex restriction enzymes, termed engineered nucleases, that are capable of making double-
Another recently developed genome manipulation technique uses CRISPR RNAs, which are small RNAs found in bacteria and archaea (see Chapter 10). The CRISPR RNAs are coupled with nucleases that cut DNA. The CRISPR RNA binds to a specific DNA sequence, and its associated nuclease then cleaves the DNA at the recognized sequence.  TRY PROBLEM 19
TRY PROBLEM 19
CONCEPTS
Restriction enzymes cut DNA at specific base sequences that are palindromic. Some restriction enzymes make staggered cuts, producing DNA fragments with cohesive ends; others cut both strands straight across, producing blunt-
 CONCEPT CHECK 1
CONCEPT CHECK 1
Where do restriction enzymes come from?
Restriction enzymes exist naturally in bacteria, which use them in defense against viruses.