Constructing a Genetic Map with Two-
Genetic maps can be constructed by conducting a series of testcrosses. In each testcross, one of the parents is heterozygous for a different pair of genes, and recombination frequencies are calculated between pairs of genes. A testcross between two genes is called a two-
129
| Gene loci in testcross | Recombination frequency (%) |
|---|---|
| a and b | 50 |
| a and c | 50 |
| a and d | 50 |
| b and c | 20 |
| b and d | 10 |
| c and d | 28 |
We can begin constructing a genetic map for these genes by considering the recombination frequencies for each pair of genes. The recombination frequency between a and b is 50%, which is the recombination frequency expected with independent assortment. Therefore, genes a and b may either be on different chromosomes or very far apart on the same chromosome; we will place them in different linkage groups with the understanding that they may or may not be on the same chromosome:
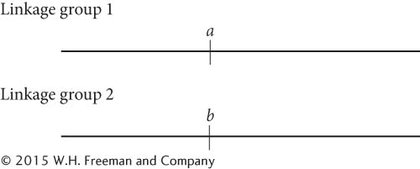
The recombination frequency between a and c is 50%, indicating that they, too, are in different linkage groups. The recombination frequency between b and c is 20%, so these genes are linked and separated by 20 map units:
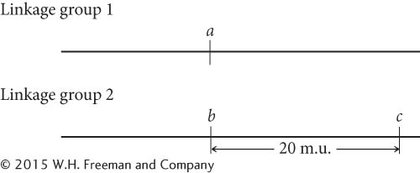
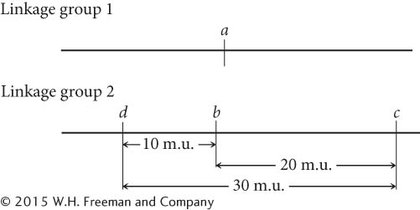
The recombination frequency between a and d is 50%, indicating that these genes belong to different linkage groups, whereas genes b and d are linked, with a recombination frequency of 10%. To decide whether gene d is 10 m.u. to the left or to the right of gene b, we must consult the c-
By examining the recombination frequency between c and d, we can distinguish between these two possibilities. The recombination frequency between c and d is 28%, so gene d must lie to the left of gene b. Notice that the sum of the recombination frequency between d and b (10%) and between b and c (20%) is greater than the recombination frequency between d and c (28%). As already discussed, this discrepancy arises because double crossovers between the two outer genes go undetected, causing an underestimation of the true map distance. The genetic map of these genes is now complete:
 TRY PROBLEM 18
TRY PROBLEM 18