Chapter 33
A nucleoside is a base attached to a ribose sugar. A nucleotide is a nucleoside with one or more phosphoryl groups attached to the ribose.
Hydrogen-
bond pairing between the base A and the base T as well as hydrogen- bond pairing between the base G and the base C in DNA. Complete the interactive matching exercise to see answers.
T is always equal to A, and so these two nucleotides constitute 40% of the bases. G is always equal to C, and so the remaining 60% must be 30% G and 30% C.
Nothing, because the base-
pair rules do not apply to single- stranded nucleic acids. Two purines are too large to fit inside the double helix, and two pyrimidines are too small to form base pairs with each other.
(a) TTGATC; (b) GTTCGA; (c) ACGCGT; (d) ATGGTA
(a) [T] + [C] = 0.46; (b) [T] = 0.30, [C] = 0.24, and [A] + [G] = 0.46
The heterocyclic bases readily absorb ultraviolet lights. The bases in the interior of a double helix are stacked and not as exposed as the bases in single-
stranded DNA. Consequently, the bases of double- stranded DNA absorb light less efficiently than single- stranded DNA. The diameter of DNA is 20 Å and 1 Å = 0.1 nm, so the diameter is 2 nm. Because 1 µm = 103 nm, the length is 2 × 104 nm. Thus, the axial ratio is 1 × 104.
The thermal energy causes the strands to wiggle about, which disrupts the hydrogen bonds between base pairs and the stacking forces between bases and thereby causes the strands to separate.
One end of a nucleic acid polymer ends with a free 5′-hydroxyl group (or a phosphoryl group esterified to the hydroxyl group), and the other end has a free 3′-hydroxyl group. Thus, the ends are different. Two strands of DNA can form a double helix only if the strands are running in different directions—
that is, have opposite directionality. 5.88 × 103 base pairs
C35
In conservative replication, after 1.0 generation, half of the molecules would be 15N-
15N and the other half would be 14N- 14N. After 2.0 generations, one- quarter of the molecules would be 15N- 15N and the other three- quarters would be 14N- 14N. Hybrid 14N- 15N molecules would not be observed in conservative replication. There would be too much charge repulsion from the negative charges on the phosphoryl groups. These charges must be countered by the addition of cations.
The nucleosome structure results in a packaging ratio of only 7. Other structures—
such as the 30- nm chromatin fiber— must form to attain the compaction ratio of 104 found in metaphase chromosomes. The DNA is bound to the histones. Consequently, it is not readily accessible to DNase. However, with an extended reaction time or more enzyme, the DNA becomes available for digestion.
The distribution of charged amino acids is H2A (13 K, 13 R, 2 D, 7 E, charge = +15), H2B (20 K, 8 R, 3 D, 7 E, charge = +18), H3 (13 K, 18 R, 4 D, 7 E, charge = +20), H4 (11 K, 14 R, 3 D, 4 E, charge = +18). The total charge of the histone octamer is estimated to be 2 × (15 + 18 + 20 + 18) = +142. The total charge on 150 base pairs of DNA is −300. Thus, the histone octamer neutralizes approximately one-
half of the charge. The total length of the DNA is estimated to be 145 bp × 3.4 Å/bp = 493 Å, which represents 1.75 turns. Thus, the circumference is 281 Å. The formula for the circumference is C = 2πr. Solving for r yields 41 Å.
GC base pairs have three hydrogen bonds compared with two for AT base pairs. Thus, the higher content of GC means more hydrogen bonds and greater helix stability.
C0t value essentially corresponds to the complexity of the DNA sequence—
in other words, how long it will take for a sequence of DNA to find its complementary strand to form a double helix. Simpler or small DNA sequences will more rapidly reanneal and have lower C0t values. Increasing amounts of salt increase the melting temperature. Because the DNA backbone is negatively charged, there is a tendency for charge repulsion to destabilize the helix and cause it to melt. The addition of salt neutralizes the charge repulsion, thereby stabilizing the double helix. The results show that, within the parameters of the experiment, more salt results in more stabilization, which gives the DNA a higher melting temperature.
The probability that any sequence will appear is 1/4n, where 4 is the number of nucleotides and n is the length of the sequence. The probability of any 15-
base sequence appearing is 1/415, or 1/1,073,741,824. Thus, a 15- nucleotide sequence would be likely to appear approximately three times (3 billion × probability of appearance). The probability of a 16- base sequence appearing is 1/416, which is equal to 1/4,294,967,296. Such a sequence will be unlikely to appear more than once. 48 = 65,536. In computer terminology, there are 64K 8-
mers of DNA. A bit specifies two bases (say, A and C) and a second bit specifies the other two (G and T). Hence, two bits are needed to specify a single nucleotide (base pair) in DNA. For example, 00, 01, 10, and 11 could encode A, C, G, and T. An 8-
mer stores 16 bits (216 = 65,536), the E. coli genome (4.6 × 106 bp) stores 9.2 × 106 bits, and the human genome (3.0 × 109 bases) stores 6.0 × 109 bits of genetic information. A 700 megabyte CD is equal to 5.6 × 109 bits. A large number of 8-
mer sequences could be stored on such a CD. The DNA sequence of E. coli could be written on a single CD with room to spare for a lot of music. One CD would not be quite enough to record the entire human genome.
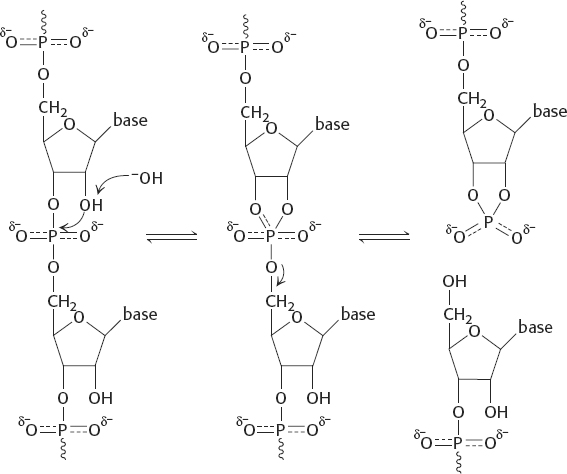
The 2′-OH group in RNA acts as an intramolecular nucleophile. In the alkaline hydrolysis of RNA, it forms a 2′-3′cyclic intermediate.