6.4Evolutionary Trees Can Be Constructed on the Basis of Sequence Information
Evolutionary Trees Can Be Constructed on the Basis of Sequence Information
The observation that homology is often manifested as sequence similarity suggests that the evolutionary pathway relating the members of a family of proteins may be deduced by examination of sequence similarity. This approach is based on the notion that sequences that are more similar to one another have had less evolutionary time to diverge than have sequences that are less similar. This method can be illustrated by using the three globin sequences in Figures 6.11 and 6.13, as well as the sequence for the human hemoglobin β chain. These sequences can be aligned with the additional constraint that gaps, if present, should be at the same positions in all of the proteins. These aligned sequences can be used to construct an evolutionary tree in which the length of the branch connecting each pair of proteins is proportional to the number of amino acid differences between the sequences (Figure 6.21).
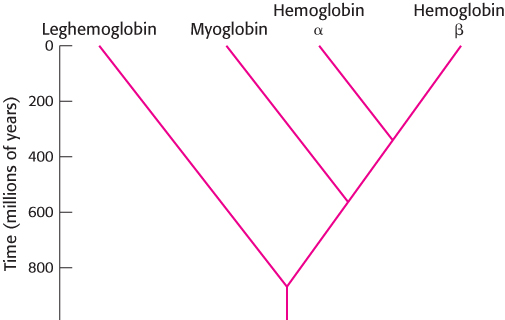

Such comparisons reveal only the relative divergence times—
184
Horizontal gene transfer events may explain unexpected branches of the evolutionary tree
Evolutionary trees that encompass orthologs of a particular protein across a range of species can lead to unexpected findings. For example, let us consider the unicellular red alga Galdieria sulphuraria, a remarkable eukaryote that can thrive in extreme environments, including at temperatures up to 56°C, at pH values between 0 and 4, and in the presence of high concentrations of toxic metals. G. sulphuraria belongs to the order Cyanidiales, clearly within the eukaryotic branch of the evolutionary tree (Figure 6.23A). However, the complete genome sequence of this organism revealed that nearly 5% of the G. sulphuraria ORFs encode proteins that are more closely related to bacterial or archaeal, not eukaryotic, orthologs. Furthermore, the proteins that exhibited these unexpected evolutionary relationships possess functions that are likely to confer a survival advantage in extreme environments, such as the removal of metal ions from inside the cell (Figure 6.23B). One likely explanation for these observations is horizontal gene transfer, or the exchange of DNA between species that provides a selective advantage to the recipient. Amongst prokaryotes, horizontal gene transfer is a well-
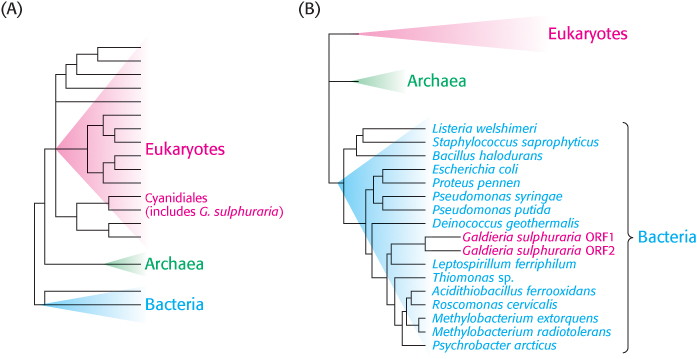
185