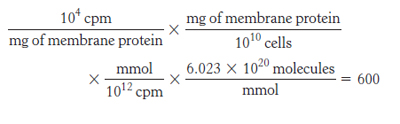Chapter 14
Chapter 14
1. The negatively charged glutamate residues mimic the negatively charged phosphoserine or phosphothreonine residues and stabilize the active conformation of the enzyme.
2. No. Phosphoserine and phosphothreonine are considerably shorter than phosphotyrosine.
3. The GTPase activity terminates the signal. Without such activity, after a pathway has been activated, it remains activated and is unresponsive to changes in the initial signal. If the GTPase activity were more efficient, the lifetime of the GTP-
4. Two identical receptor molecules must recognize different aspects of the same signal molecule.
5. Growth-
6. The mutated α subunit will always be in the GTP form and, hence, in the active form, which would stimulate its signaling pathway.
7. A G protein is a component of the signal-
8. Calcium ions diffuse slowly because they bind to many protein surfaces within a cell, impeding their free motion. Cyclic AMP does not bind as frequently, and so it diffuses more rapidly.
9. Fura-
10. Gαs stimulates adenylate cyclase, leading to the generation of cAMP. This signal then leads to glucose mobilization (Chapter 21). If cAMP phosphodiesterase were inhibited, then cAMP levels would remain high even after the termination of the epinephrine signal, and glucose mobilization would continue.
11. If the two kinase domains are forced to be within close proximity of each other, the activation loop of one kinase, in its inactivating conformation, can be displaced by the activation loop of the neighboring kinase, which acts as a substrate for phosphorylation.
12. The full network of pathways initiated by insulin includes a large number of proteins and is substantially more elaborate than indicated in Figure 14.26. Furthermore, many additional proteins take part in the termination of insulin signaling. A defect in any of the proteins in the insulin signaling pathways or in the subsequent termination of the insulin response could potentially cause problems. Therefore, it is not surprising that many different gene defects can cause type 2 diabetes.
13. The binding of growth hormone causes its monomeric receptor to dimerize. The dimeric receptor can then activate a separate tyrosine kinase to which the receptor binds. The signaling pathway can then continue in similar fashion to the pathways that are activated by the insulin receptor or other mammalian EGF receptors.
14. The truncated receptor will dimerize with the full-
15. Insulin would elicit the response that is normally caused by EGF. Insulin binding will likely stimulate dimerization and phosphorylation of the chimeric receptor and thereby signal the downstream events that are normally triggered by EGF binding. Exposure of these cells to EGF would have no effect.
16. 105
17. The formation of diacylglycerol implies the participation of phospholipase C. A simple pathway would entail receptor activation by cross-
A18
18. Other potential drug targets within the EGF signaling cascade include, but are not limited to, the kinase active sites of the EGF receptor, Raf, MEK, or ERK.
19. In the reaction catalyzed by adenylate cyclase, the 3′-OH group nucleophilically attacks the α-phosphorus atom attached to the 5′-OH group, leading to displacement of pyrophosphate. The reaction catalyzed by DNA polymerase is similar except that the 3′-OH group is on a different nucleotide.
20. ATP-
21. (a) X ≈ 10−7 M; Y ≈ 5 × 10−6 M; Z ≈ 10−3 M. (b) Because much less X is required to fill half of the sites, X displays the highest affinity. (c) The binding affinity almost perfectly matches the ability to stimulate adenylate cyclase, suggesting that the hormone–
22. (a) The total binding does not distinguish binding to a specific receptor from binding to different receptors or from nonspecific binding to the membrane.
(b) The rationale is that the receptor will have a high affinity for the ligand. Thus, in the presence of excess nonradioactive ligand, the receptor will bind to nonradioactive ligand. Therefore, any binding of the radioactive ligand must be nonspecific.
(c) The plateau suggests that the number of receptor-
23. Number of receptors per cell =