25.1The Pyrimidine Ring Is Assembled de Novo or Recovered by Salvage Pathways
The Pyrimidine Ring Is Assembled de Novo or Recovered by Salvage Pathways
In de novo synthesis of pyrimidines, the ring is synthesized first and then it is attached to a ribose phosphate to form a pyrimidine nucleotide (Figure 25.2). Pyrimidine rings are assembled from bicarbonate, aspartate, and ammonia. Although an ammonia molecule already present in solution can be used, the ammonia is usually produced from the hydrolysis of the side chain of glutamine.
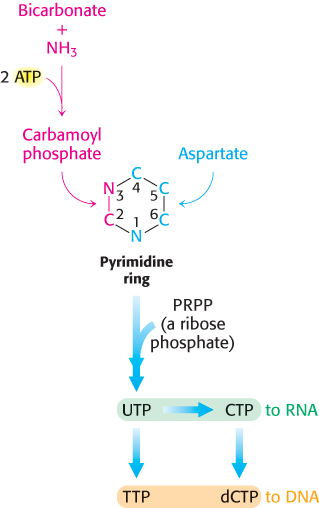
745
Bicarbonate and other oxygenated carbon compounds are activated by phosphorylation
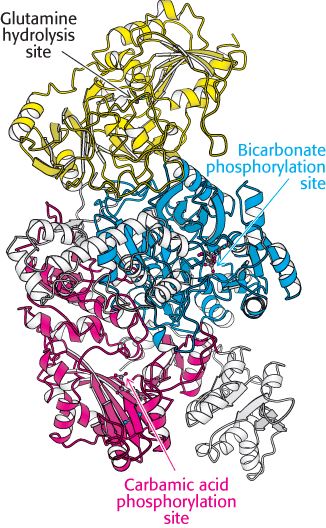
 FIGURE 25.3 Structure of carbamoyl phosphate synthetase II. Notice that the enzyme contains sites for three reactions. This enzyme consists of two chains. The smaller chain (yellow) contains a site for glutamine hydrolysis to generate ammonia. The larger chain includes two ATP-
FIGURE 25.3 Structure of carbamoyl phosphate synthetase II. Notice that the enzyme contains sites for three reactions. This enzyme consists of two chains. The smaller chain (yellow) contains a site for glutamine hydrolysis to generate ammonia. The larger chain includes two ATP-The first step in de novo pyrimidine biosynthesis is the synthesis of carbamoyl phosphate from bicarbonate and ammonia in a multistep process, requiring the cleavage of two molecules of ATP. This reaction is catalyzed by carbamoyl phosphate synthetase II (CPS II). Recall that carbamoyl phosphate synthetase I facilitates ammonia incorporation into urea (Section 23.4). Carbamoyl phosphate synthetase II is a dimer composed of a small subunit that hydrolyzes glutamine to form NH3 and a large subunit that completes the synthesis of carbamoyl phosphate. Analysis of the structure of the large subunit reveals two homologous domains, each of which catalyzes an ATP-
In the first step, bicarbonate is phosphorylated by ATP to form carboxyphosphate and ADP. Ammonia then reacts with carboxyphosphate to form carbamic acid and inorganic phosphate.
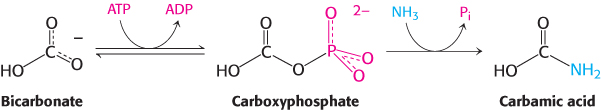
The active site for this reaction lies in a domain formed by the amino-
In the second step catalyzed by carbamoyl phosphate synthetase II, carbamic acid is phosphorylated by another molecule of ATP to form carbamoyl phosphate.
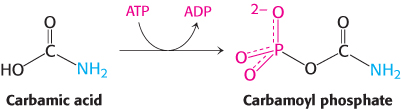
This reaction takes place in a second ATP-
The side chain of glutamine can be hydrolyzed to generate ammonia
Glutamine is the primary source of ammonia for carbamoyl phosphate synthetase II. In this case, the small subunit of the enzyme hydrolyzes glutamine to form ammonia and glutamate. The active site of the glutamine-
Intermediates can move between active sites by channeling
Carbamoyl phosphate synthetase II contains three different active sites (Figure 25.3), separated from one another by a total of 80 Å. Intermediates generated at one site move to the next without leaving the enzyme. These intermediates move within the enzyme by means of substrate channeling, similar to the process described for tryptophan synthetase (Figure 25.4; also Figure 24.16). The ammonia generated in the glutamine-
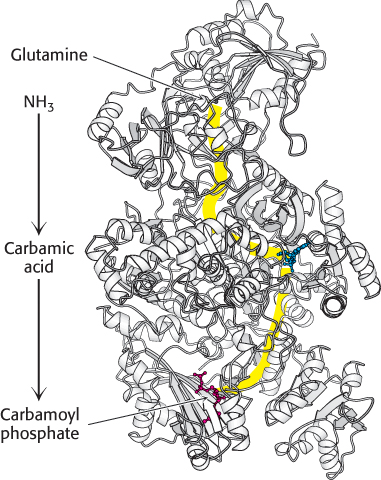
 FIGURE 25.4 Substrate channeling. The three active sites of carbamoyl phosphate synthetase II are linked by a channel (yellow) through which intermediates pass. Glutamine enters one active site, and carbamoyl phosphate, which includes the nitrogen atom from the glutamine side chain, leaves another 80 Å away.
FIGURE 25.4 Substrate channeling. The three active sites of carbamoyl phosphate synthetase II are linked by a channel (yellow) through which intermediates pass. Glutamine enters one active site, and carbamoyl phosphate, which includes the nitrogen atom from the glutamine side chain, leaves another 80 Å away.
746
Orotate acquires a ribose ring from PRPP to form a pyrimidine nucleotide and is converted into uridylate
Carbamoyl phosphate reacts with aspartate to form carbamoylaspartate in a reaction catalyzed by aspartate transcarbamoylase (Section 10.1). Carbamoylaspartate then cyclizes to form dihydroorotate, which is then oxidized by NAD+ to form orotate.
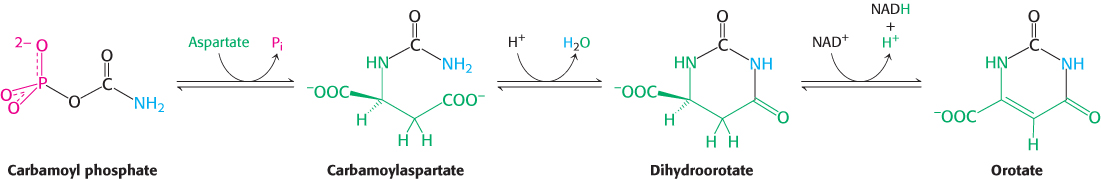
In mammals, the enzymes that form orotate are part of single large polypeptide chain called CAD, for carbamoyl phosphate synthetase, aspartate transcarbamoylase and dihydroorotase.
At this stage, orotate couples to ribose, in the form of 5-
747
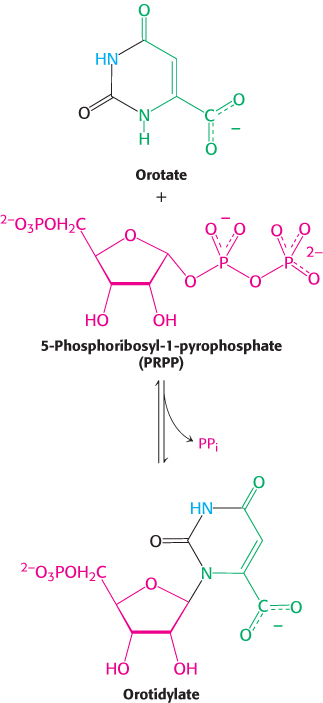
Orotate reacts with PRPP to form orotidylate, a pyrimidine nucleotide. This reaction is driven by the hydrolysis of pyrophosphate. The enzyme that catalyzes this addition, orotate phosphoribosyltransferase, is homologous to a number of other phosphoribosyltransferases that add different groups to PRPP to form the other nucleotides. Orotidylate is then decarboxylated to form uridylate (UMP), a major pyrimidine nucleotide that is a precursor to RNA. This reaction is catalyzed by orotidylate decarboxylase, also called orotidine-
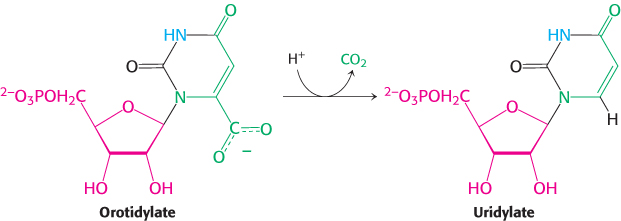
Orotidylate decarboxylase is one of the most proficient enzymes known. In its absence, decarboxylation is extremely slow and is estimated to take place once every 78 million years; with the enzyme present, it takes place approximately once per second, a rate enhancement of 1017-fold. The phosphoribosyltransferase and decarboxylase activities are located on the same polypeptide chain, providing another example of a bifunctional enzyme. The bifunctional enzyme is called uridine monophosphate synthetase.
Nucleotide mono-, di-, and triphosphates are interconvertible
How is the other major pyrimidine ribonucleotide, cytidine, formed? It is synthesized from the uracil base of UMP, but the synthesis can take place only after UMP has been converted into UTP. Recall that the diphosphates and triphosphates are the active forms of nucleotides in biosynthesis and energy conversions. Nucleoside monophosphates are converted into nucleoside triphosphates in stages. First, nucleoside monophosphates are converted into diphosphates by specific nucleoside monophosphate kinases that utilize ATP as the phosphoryl-

Nucleoside diphosphates and triphosphates are interconverted by nucleoside diphosphate kinase, an enzyme that has broad specificity, in contrast with the monophosphate kinases. X and Y represent any of several ribonucleosides or even deoxyribonucleosides:

CTP is formed by amination of UTP
After uridine triphosphate has been formed, it can be transformed into cytidine triphosphate by the replacement of a carbonyl group by an amino group, a reaction catalyzed by cytidine triphosphate synthetase.
748
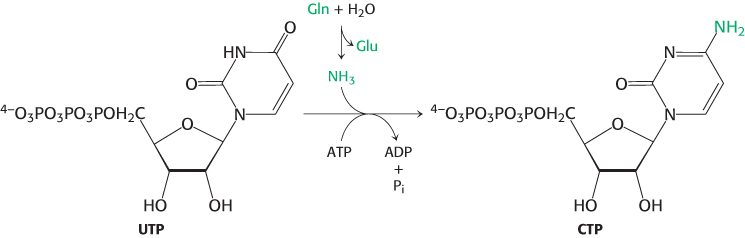
Like the synthesis of carbamoyl phosphate, this reaction requires ATP and uses glutamine as the source of the amino group. The reaction proceeds through an analogous mechanism in which the O-
Salvage pathways recycle pyrimidine bases
Pyrimidine bases can be recovered from the breakdown products of DNA and RNA by the use of salvage pathways. In these pathways, a preformed base is reincorporated into a nucleotide. We will consider the salvage for the pyrimidine base thymine. Thymine is found in DNA and base-
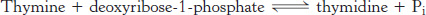
Thymidine is then converted into a nucleotide by thymidine kinase.
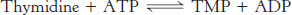
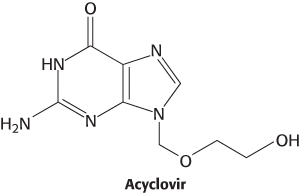
 Viral thymidine kinase differs from the mammalian enzyme and thus provides a therapeutic target. For instance, herpes simplex infections are treated with acyclovir (acycloguanosine), which viral thymidine kinase converts into acyclovir monophosphate by adding a phosphate to the hydroxyl group of acyclovir. Viral thymidine kinase binds acyclovir more than 200 times as tightly as cellular thymidine kinase, accounting for its effect on infected cells only. Acyclovir monophosphate is phosphorylated by cellular enzymes to yield acyclovir triphosphate. Acyclovir triphosphate competes with dGTP for DNA polymerase. Once incorporated into viral DNA, it acts as a chain terminator since it lacks a 3′-hydroxyl required for chain extension. As we will see shortly, thymidine kinase also plays a role in the de novo synthesis of thymidylate.
Viral thymidine kinase differs from the mammalian enzyme and thus provides a therapeutic target. For instance, herpes simplex infections are treated with acyclovir (acycloguanosine), which viral thymidine kinase converts into acyclovir monophosphate by adding a phosphate to the hydroxyl group of acyclovir. Viral thymidine kinase binds acyclovir more than 200 times as tightly as cellular thymidine kinase, accounting for its effect on infected cells only. Acyclovir monophosphate is phosphorylated by cellular enzymes to yield acyclovir triphosphate. Acyclovir triphosphate competes with dGTP for DNA polymerase. Once incorporated into viral DNA, it acts as a chain terminator since it lacks a 3′-hydroxyl required for chain extension. As we will see shortly, thymidine kinase also plays a role in the de novo synthesis of thymidylate.