31.4Gene Expression Can Be Controlled at Posttranscriptional Levels
Gene Expression Can Be Controlled at Posttranscriptional Levels
The modulation of the rate of transcription initiation is the most common mechanism of gene regulation. However, other stages of transcription also can be targets for regulation. In addition, the process of translation provides other points of intervention for regulating the level of a protein produced in a cell. In Chapter 29, we considered riboswitches that control transcription termination (Section 29.1). Other riboswitches control gene expression by other mechanisms such as the formation of structures that inhibit translation. Additional mechanisms for posttranscriptional gene regulation have been discovered, one of which will be described here.
Attenuation is a prokaryotic mechanism for regulating transcription through the modulation of nascent RNA secondary structure
A means for regulating transcription in bacteria was discovered by Charles Yanofsky and his colleagues as a result of their studies of the tryptophan operon. This operon encodes five enzymes that convert chorismate into tryptophan. Analysis of the 5′ end of trp mRNA revealed the presence of a leader sequence of 162 nucleotides before the initiation codon of the first enzyme. The next striking observation was that bacteria produced a transcript consisting of only the first 130 nucleotides when the tryptophan level was high, but they produced a 7000-
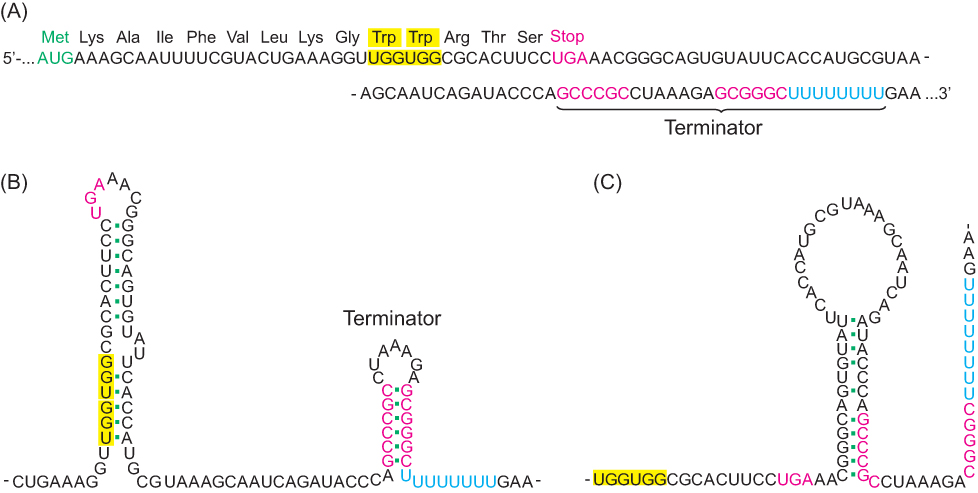
Attenuation depends on features at the 5′ end of the mRNA product (Figure 31.20). The first part of the leader sequence encodes a 14-
936
How does the level of tryptophan alter transcription of the trp operon? An important clue was the finding that the 14-
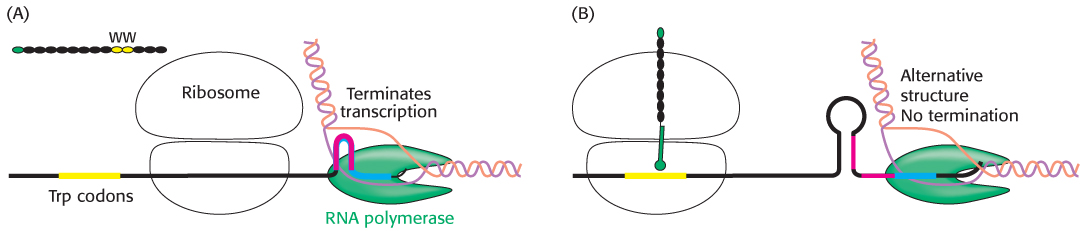
Several other operons for the biosynthesis of amino acids in E. coli also are regulated by attenuator sites. The leader peptide of each contains an abundance of the amino acid residues of the type synthesized by the operon (Figure 31.22). For example, the leader peptide for the phenylalanine operon includes 7 phenylalanine residues among 15 residues. The threonine operon encodes enzymes required for the synthesis of both threonine and isoleucine; the leader peptide contains 8 threonine and 4 isoleucine residues in a 16-

937
 The examples of prokaryotic gene regulation that we have been describing come from bacteria, as opposed to archaea. The transcriptional apparatus present in archaea shares many features with that found in eukaryotes. This commonality is frequently interpreted to suggest that eukaryotes evolved after a cell fusion event in which a bacterial cell was engulfed by an archaeal cell. Nonetheless, the key principles of gene regulation such as the occurrence of operons and the roles of DNA-
The examples of prokaryotic gene regulation that we have been describing come from bacteria, as opposed to archaea. The transcriptional apparatus present in archaea shares many features with that found in eukaryotes. This commonality is frequently interpreted to suggest that eukaryotes evolved after a cell fusion event in which a bacterial cell was engulfed by an archaeal cell. Nonetheless, the key principles of gene regulation such as the occurrence of operons and the roles of DNA-