32.4
Eukaryotic Gene Expression Can Be Controlled at Posttranscriptional Levels
Just as in prokaryotes, gene expression in eukaryotes can be regulated subsequent to transcription. We shall consider two examples. The first is the regulation of genes taking part in iron metabolism through key features in RNA secondary structure, similar in many ways to prokaryotic posttranscriptional regulation (Section 31.4). The second entails an entirely new mechanism, first glimpsed with the discovery of RNA interference. Certain small regulatory RNA molecules allow the regulation of gene expression through interaction with a range of mRNA molecules. Remarkably, this mechanism, discovered only recently, affects the expression of approximately 60% of all human genes.
Genes associated with iron metabolism are translationally regulated in animals
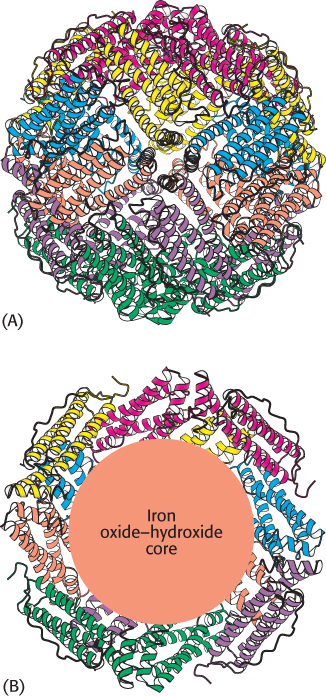
 FIGURE 32.22 Structure of ferritin. (A) Twenty-four ferritin polypeptides form a nearly spherical shell. (B) A cutaway view reveals the core that stores iron as an iron oxide–hydroxide complex
FIGURE 32.22 Structure of ferritin. (A) Twenty-four ferritin polypeptides form a nearly spherical shell. (B) A cutaway view reveals the core that stores iron as an iron oxide–hydroxide complex
[Drawn from 1IES.pdb.]
RNA secondary structure plays a role in the regulation of iron metabolism in eukaryotes. Iron is an essential nutrient, required for the synthesis of hemoglobin, cytochromes, and many other proteins. However, excess iron can be quite harmful because, untamed by a suitable protein environment, iron can initiate a range of free-radical reactions that damage proteins, lipids, and nucleic acids. Animals have evolved sophisticated systems for the accumulation of iron in times of scarcity and for the safe storage of excess iron for later use. Key proteins include transferrin, a transport protein that carries iron in the serum, transferrin receptor, a membrane protein that binds iron-loaded transferrin and initiates its entry into cells, and ferritin, an impressively efficient iron-storage protein found primarily in the liver and kidneys. Twenty-four ferritin polypeptides form a nearly spherical shell that encloses as many as 2400 iron atoms, a ratio of one iron atom per amino acid (Figure 32.22).
Ferritin and transferrin-receptor expression levels are reciprocally related in their responses to changes in iron levels. When iron is scarce, the amount of transferrin receptor increases and little or no new ferritin is synthesized. Interestingly, the extent of mRNA synthesis for these proteins does not change correspondingly. Instead, regulation takes place at the level of translation.
Consider ferritin first. Ferritin mRNA includes a stem-loop structure termed an iron-response element (IRE) in its 5′ untranslated region (Figure 32.23). This stem-loop binds a 90-kDa protein, called an IRE-binding protein (IRP), that blocks the initiation of translation as the IRE binds on the 5′ side of the coding region. When the iron level increases, the IRP binds iron as a 4Fe-4S cluster. The IRP bound to iron cannot bind RNA, because the binding sites for iron and RNA substantially overlap. Thus, in the presence of iron, ferritin mRNA is released from the IRP and translated to produce ferritin, which sequesters the excess iron.
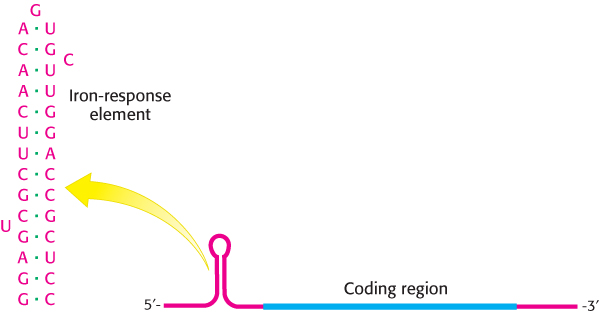
FIGURE 32.23Iron-response element. Ferritin mRNA includes a stem-loop structure, termed an iron-response element (IRE), in its 5′ untranslated region. The IRE binds a specific protein that blocks the translation of this mRNA under low iron conditions.
An examination of the nucleotide sequence of transferrin-receptor mRNA reveals the presence of several IRE-like regions. However, these regions are located in the 3′ untranslated region rather than in the 5′ untranslated region (Figure 32.24). Under low-iron conditions, IRP binds to these IREs. However, given the location of these binding sites, the transferrin-receptor mRNA can still be translated. What happens when the iron level increases and the IRP no longer binds transferrin-receptor mRNA? Freed from the IRP, transferrin-receptor mRNA is rapidly degraded. Thus, an increase in the cellular iron level leads to the destruction of transferrin-receptor mRNA and, hence, a reduction in the production of transferrin-receptor protein.
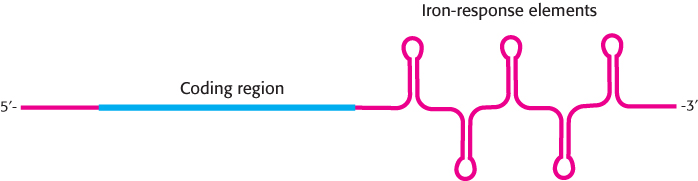
FIGURE 32.24Transferrin-receptor mRNA. This mRNA has a set of iron-response elements (IREs) in its 3′ untranslated region. The binding of the IRE-binding protein to these elements stabilizes the mRNA but does not interfere with translation.
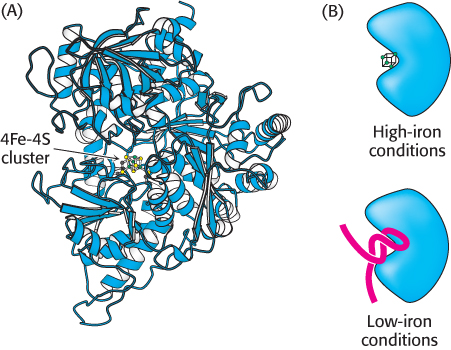
 FIGURE 32.25 The IRP is an aconitase. (A) Aconitase contains an unstable 4Fe-4S cluster at its center. (B) Under conditions of low iron, the 4Fe-4S cluster dissociates and appropriate RNA molecules can bind in its place.
FIGURE 32.25 The IRP is an aconitase. (A) Aconitase contains an unstable 4Fe-4S cluster at its center. (B) Under conditions of low iron, the 4Fe-4S cluster dissociates and appropriate RNA molecules can bind in its place.
[Drawn from 1C96.pdb.]
 The purification of the IRP and the cloning of its cDNA were sources of truly remarkable insight into evolution. The IRP was found to be approximately 30% identical in amino acid sequence with the citric acid cycle enzyme aconitase from mitochondria. Further analysis revealed that the IRP is, in fact, an active aconitase enzyme; it is a cytoplasmic aconitase that had been known for a long time, but its function was not well understood (Figure 32.25). The iron–sulfur center at the active site of the IRP is rather unstable, and loss of the iron triggers significant changes in protein conformation. Thus, this protein can serve as an iron-sensing factor.
The purification of the IRP and the cloning of its cDNA were sources of truly remarkable insight into evolution. The IRP was found to be approximately 30% identical in amino acid sequence with the citric acid cycle enzyme aconitase from mitochondria. Further analysis revealed that the IRP is, in fact, an active aconitase enzyme; it is a cytoplasmic aconitase that had been known for a long time, but its function was not well understood (Figure 32.25). The iron–sulfur center at the active site of the IRP is rather unstable, and loss of the iron triggers significant changes in protein conformation. Thus, this protein can serve as an iron-sensing factor.
Other mRNAs, including those taking part in heme synthesis, have been found to contain IREs. Thus, genes encoding proteins required for iron metabolism acquired sequences that, when transcribed, provided binding sites for the iron-sensing protein. An environmental signal—the concentration of iron—controls the translation of proteins required for the metabolism of this metal. Thus, mutations in the untranslated region of mRNAs have been selected for beneficial regulation by iron levels.
Small RNAs regulate the expression of many eukaryotic genes
Genetic studies of development in C. elegans revealed that a gene called lin-4 encodes an RNA molecule 61 nucleotides long that can regulate the expression of certain other genes. The 61-nucleotide lin-4 RNA does not encode a protein, but rather is cleaved into a 22-nucleotide RNA that possesses the regulatory activity. This discovery was the first view of a large class of regulatory RNA molecules that are now called microRNAs or miRNAs. The key to the activity of microRNAs and their specificity for particular genes is their ability to form Watson–Crick base-pair-stabilized complexes with the mRNAs of those genes.
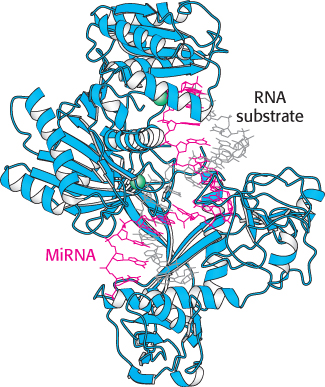
 FIGURE 32.26 MicroRNA–Argonaute complex. The miRNA (shown in red) is bound by the Argonaute protein. Notice that the miRNA serves as a guide that binds the RNA substrate (shown in gray) through the formation of a double helix. Two magnesium ions are shown in green.
FIGURE 32.26 MicroRNA–Argonaute complex. The miRNA (shown in red) is bound by the Argonaute protein. Notice that the miRNA serves as a guide that binds the RNA substrate (shown in gray) through the formation of a double helix. Two magnesium ions are shown in green.
[Drawn from 3HK2.pdb.]
These miRNAs do not function on their own. Instead, the miRNAs bind to members of a class of proteins called the Argonaute family (Figure 32.26). These Argonaute–miRNA complexes can then bind mRNAs that have sequences that are substantially complementary to the miRNAs. Once bound, such an mRNA can be cleaved by the Argonaute–miRNA complex through the action of a magnesium-based active site. Thus, the miRNAs serve as guide RNAs that determine the specificity of the Argonaute complex (Figure 32.27). Cleavage of mRNA by the Argonaute–miRNA complex is similar to the mechanism of RNA interference. The small RNA molecules participating in RNAi, however, come from a different source. In RNAi, double-stranded RNAs are cleaved into 21-nucleotide fragments, single-stranded components of which are bound by members of the Argonaute family to form an RNA-induced silencing complex (RISC) that cleaves complementary mRNAs. For miRNAs, the single-stranded RNAs are generated from larger genetically encoded precursors, as described in Chapter 29.
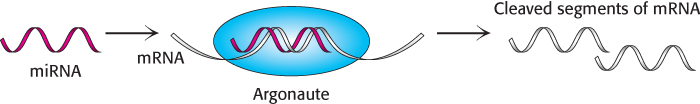
FIGURE 32.27MicroRNA action. MicroRNAs bind to members of the Argonaute family where they target specific mRNA molecules for cleavage.
The occurrence of gene regulation by miRNAs was originally thought to be limited to a relatively small number of species. However, subsequent studies have revealed that this mode of gene regulation is nearly ubiquitous in eukaryotes. Indeed, more than 700 miRNAs encoded by the human genome have been identified. Each miRNA can regulate many different genes because many different target sequences are present in each mRNA. An estimated 60% of all human genes are regulated by one or more miRNAs. As one example, consider the human miRNA called miR-206. This miRNA down-regulates the expression of one isoform of the estrogen receptor. In addition, it appears to down-regulate the expression of several different coactivators that interact with the estrogen receptor. Thus, this miRNA can mute the influence of estrogen by blocking the estrogen-initiated signaling pathway at several different steps.
 The microRNA pathway has tremendous implications for the evolution of gene-regulatory pathways. Most of the target sites for miRNAs are present in the 3′-untranslated regions of mRNAs. These sequences are quite free to mutate because they do not encode proteins and are not required to fold into any specific structures. Thus, in the context of a set of expressed miRNAs, mutations in this region of any particular gene could, in principle, increase or decrease the affinity for one or more miRNAs and, hence, alter the regulation of the gene.
The microRNA pathway has tremendous implications for the evolution of gene-regulatory pathways. Most of the target sites for miRNAs are present in the 3′-untranslated regions of mRNAs. These sequences are quite free to mutate because they do not encode proteins and are not required to fold into any specific structures. Thus, in the context of a set of expressed miRNAs, mutations in this region of any particular gene could, in principle, increase or decrease the affinity for one or more miRNAs and, hence, alter the regulation of the gene.

 FIGURE 32.22 Structure of ferritin. (A) Twenty-
FIGURE 32.22 Structure of ferritin. (A) Twenty-


 FIGURE 32.25 The IRP is an aconitase. (A) Aconitase contains an unstable 4Fe-
FIGURE 32.25 The IRP is an aconitase. (A) Aconitase contains an unstable 4Fe- The purification of the IRP and the cloning of its cDNA were sources of truly remarkable insight into evolution. The IRP was found to be approximately 30% identical in amino acid sequence with the citric acid cycle enzyme aconitase from mitochondria. Further analysis revealed that the IRP is, in fact, an active aconitase enzyme; it is a cytoplasmic aconitase that had been known for a long time, but its function was not well understood (Figure 32.25). The iron–
The purification of the IRP and the cloning of its cDNA were sources of truly remarkable insight into evolution. The IRP was found to be approximately 30% identical in amino acid sequence with the citric acid cycle enzyme aconitase from mitochondria. Further analysis revealed that the IRP is, in fact, an active aconitase enzyme; it is a cytoplasmic aconitase that had been known for a long time, but its function was not well understood (Figure 32.25). The iron–
 FIGURE 32.26 MicroRNA–
FIGURE 32.26 MicroRNA–
 The microRNA pathway has tremendous implications for the evolution of gene-
The microRNA pathway has tremendous implications for the evolution of gene-