PROBLEMS
PROBLEMS
Question 32.1
Charge neutralization. Given the histone amino acid sequences illustrated below, estimate the charge of a histone octamer at pH 7. Assume that histidine residues are uncharged at this pH. How does this charge compare with the charge on 150 base pairs of DNA?
Histone H2A
MSGRGKQGGKARAKAKTRSSRAGLQFPVGRVHRLLRKGNYSERVGAGAPVYLAAVLEYLTAEILELAGNA
ARDNKKTRIIPRHLQLAIRNDEELNKLLGRVTIAQGGVLPNIQAVLLPKKTESHHKAKGK
Histone H2B
MPEPAKSAPAPKKGSKKAVTKAQKKDGKKRKRSRKESYSVYVYKVLKQVHPDTGISSKAMGIMNSFVNDI
FERIAGEASRLAHYNKRSTITSREIQTAVRLLLPGELAKHAVSEGTKAVTKYTSSK
Histone H3
MARTKQTARKSTGGKAPRKQLATKAARKSAPSTGGVKKPHRYRPGTVALREIRRYQKSTELLIRKLPFQR
LVREIAQDFKTDLRFQSAAIGALQEASEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA
Histone H4
MSGRGKGGKGLGKGGAKRHRKVLRDNIQGITKPAIRRLARRGGVKRISGLIYEETRGVLKVFLENVIRDA
VTYTEHAKRKTVTAMDVVYALKRQGRTLYGFGG
Question 32.2
Chromatin immunoprecipitation. You have used the technique of chromatin immunoprecipitation to isolate DNA fragments containing a DNA-
Question 32.3
Around we go. Assuming that 145 base pairs of DNA wrap around the histone octamer 1.75 times, estimate the radius of the histone octamer. Assume 3.4 Å per base pair and simplify the calculation by assuming that the wrapping is in two rather than three dimensions and neglecting the thickness of the DNA.
Question 32.4
Nitrogen substitution. Growth of mammalian cells in the presence of 5-
959
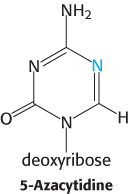
Question 32.5
A new domain. A protein domain that recognizes 5-
Question 32.6
Hybrid receptor. Through recombinant DNA methods, a modified steroid hormone receptor was prepared that consists of an estrogen receptor with its ligand-
Question 32.7
Different modifications. What is the effect of acetylation of a lysine residue on the charge of a histone protein? Of lysine methylation?
Question 32.8
Transformer. The following amino acid sequence of one of the four transcription factors is used to generate iPS cells:
HTCDYAGCGKTYTKSSHLKAHLRTHTGEKPYHCDWDGCGWKFARSDELTRHYRKHTGHRPFQCQKCD
RAFSRSDHLALHMKRHF
This transcription factor belongs to one of the three structural classes discussed in Section 32.2. Identify the class.
Question 32.9
Coverage. What percentage of the DNA sites in yeast are accessible, assuming that the fraction of sites observed for GAL4 is typical? To how many base pairs of the 12-
Question 32.10
Count the methyl groups. Examination of the histone modifications of a gene reveals an abundance of histone H3 with lysine 27 modified with a single methyl group. Does this suggest that this gene is activated or repressed? How would your answer change if many lysine 27 residues were modified with three methyl groups each?
Question 32.11
Iron regulation. What effect would you expect from the addition of an IRE to the 5′ end of a gene that is not normally regulated by iron levels? To the 3′ end?
Question 32.12
Predicting microRNA regulation. Suppose that you have identified an miRNA that has the sequence 5′-GCCUAGCCUUAGCAUUGAUUGG-
Mechanism Problem
Question 32.13
Acetyltransferases. Propose a mechanism for the transfer of an acetyl group from acetyl CoA to the amino group of lysine.
Data Interpretation Problem
Question 32.14
Limited restriction. The restriction enzyme HpaII is a powerful tool for analyzing DNA methylation. This enzyme cleaves sites of the form 5′-CCGG-
960