PROBLEMS
Question 7.1
When joining two or more DNA fragments, a researcher can adjust the sequence at the junction(s) in a variety of subtle ways, as seen in the following exercises.
Draw the structure of each end of the linear DNA fragments produced by an EcoRI restriction digest (include the sequences remaining from the EcoRI recognition sequence).
Draw the structure resulting from the reaction of these end sequences with a DNA polymerase and the four deoxynucleoside triphosphates.
Draw the sequence produced at the junction that arises when the two ends with the structure derived in (b) are ligated.
Draw the structure produced when the structure derived in (a) is treated with a nuclease that degrades only single-
stranded DNA. Draw the sequence of the junction produced when an end with structure (b) is ligated to an end with structure (d).
Draw the structure of the end of a linear DNA fragment produced by a PvuII restriction digest (include the sequences remaining from the PvuII recognition sequence).
Draw the sequence of the junction produced when an end with structure (b) is ligated to an end with structure (f).
Suppose you can synthesize a short duplex DNA fragment with any sequence you wish. With this synthetic fragment and any of the procedures described in (a) through (g), design a protocol that would remove an EcoRI restriction site from a DNA molecule and insert a new BamHI restriction site at approximately the same location. (See Figure 7-2.)
Design four different short, synthetic double-
stranded DNA fragments that would permit ligation of structure (a) with a DNA fragment produced by a PstI restriction digest. In one of these fragments, design the sequence so that the final junction contains the recognition sites for both EcoRI and PstI. In the second and third fragments, design the sequences so that the junction contains only the EcoRI and only the PstI recognition site, respectively. Design the sequence of the fourth fragment so that neither the EcoRI nor the PstI recognition site appears in the junction.
Question 7.2
The partial sequence of one strand of a double-
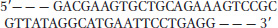
The cleavage sites for the restriction enzymes EcoRI and PstI are shown below.

Write the sequence of both strands of the DNA fragment created when this DNA is cleaved with both EcoRI and PstI. The top strand of your duplex DNA fragment should be derived from the strand sequence given above.
Question 7.3
When cloning a foreign DNA fragment into a plasmid, it is often useful to insert the fragment at a site that interrupts a selectable marker (such as the tetracycline-
Question 7.4
The plasmid cloning vector pBR322 (see Figure 7-4) is cleaved with the restriction endonuclease PstI. An isolated DNA fragment from a eukaryotic genome (also produced by PstI cleavage) is added to the prepared vector and ligated. The mixture of ligated DNAs is then used to transform bacteria, and plasmid-
254
In addition to the desired recombinant plasmid, what other types of plasmids might be found among the transformed bacteria that are tetracycline resistant? How can the types be distinguished?
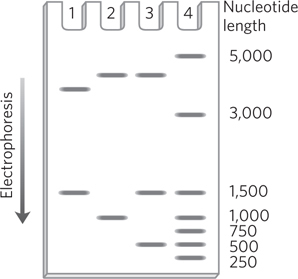
The cloned DNA fragment is 1,000 bp long and has an EcoRI site 250 bp from one end. Three different recombinant plasmids are cleaved with EcoRI and analyzed by gel electrophoresis, giving the patterns shown below. What does each pattern say about the cloned DNA? Note that in pBR322, the PstI and EcoRI restriction sites are about 750 bp apart. The entire plasmid with no cloned insert has 4,361 bp. Size markers in lane 4 have the number of nucleotides noted on the right.
Question 7.5
A new restriction endonuclease is discovered that recognizes and cleaves the palindromic sequence GGATATCC. How often does this sequence appear in a random-
Question 7.6
A BAC vector is designed so that large DNA fragments can be inserted into a cleavage site for the enzyme BamHI (see Table 7-2). To prepare chromosomal DNA from a target organism for cloning into this vector, the target DNA is treated just briefly with BamHI, not long enough to cleave all of the BamHI sites present. Explain why the BamHI reaction is halted before the chromosomal DNA is completely cleaved.
Question 7.7
One strand of a chromosomal DNA sequence is shown below. An investigator wants to amplify and isolate a DNA fragment defined by the segment shown in red, using the polymerase chain reaction. Design two PCR primers, each 20 nucleotides long, that can be used to amplify this DNA segment.
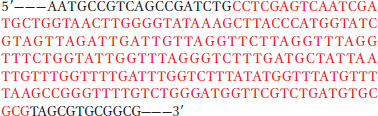
Question 7.8
A researcher wants to amplify the same DNA segment described in Problem 7. However, to aid in cloning, she wants to add a short DNA sequence on each end of the amplified segment that includes the restriction site for the enzyme EcoRI. Design the two PCR primers that this researcher needs, incorporating 20 nucleotides complementary to the appropriate target sequences.
Question 7.9
Huntington disease (HD) is an inherited neurodegenerative disorder characterized by gradual, irreversible impairment of psychological, motor, and cognitive functions. Symptoms typically appear in middle age, but onset can occur at almost any age, and the course of the disease can range from 15 to 20 years. The molecular basis of HD is becoming better understood, and the genetic mutation has been traced to a gene that encodes a protein (Mr 350,000) of unknown function. In individuals who will not develop HD, a region of the gene that encodes the N-
A small portion of the N-

Question 7.10
In a species of ciliated protist, a segment of genomic DNA is sometimes deleted. The deletion is a genetically programmed reaction associated with cellular mating. A researcher proposes that the DNA is deleted in a type of recombination called site-
255
Question 7.11
The short DNA shown below is to be sequenced. The asterisk represents a radioactive label. Using your knowledge of how the Sanger method works, in the gel diagram, draw in the bands that will appear when DNA polymerase is added to the reaction along with the four different nucleotide mixtures indicated (A through D; the bands for A are given). Note that some of these mixtures are not what would normally be used in a sequencing reaction. Dideoxynucleotides (ddNTPs) are added in relatively small amounts.
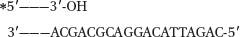
Nucleotide mixtures:
dATP, dTTP, dCTP, dGTP, ddTTP (given)
dATP, dTTP, dCTP, dGTP, ddATP
dTTP, dGTP, dCTP, ddCTP, ddATP
dATP, dCTP, dTTP, ddGTP

Question 7.12
To express a cloned gene, the DNA encoding that gene is placed downstream of a bacterial promoter. RNA polymerase binds at the promoter and moves away from it in one direction, synthesizing RNA, using one strand of the DNA as a template. The synthesized RNA strand (mRNA) carries the information specifying a protein to the ribosome. The RNA strand is synthesized in the 5′→3′ direction. The mRNA is identical in sequence (with U replacing T) to one of the DNA strands, and complementary to the other strand. If the orientation of the cloned gene is inverted relative to the promoter, will the same protein still be expressed? Why or why not?
Question 7.13
An investigator has two systems available for the cloning and expression of proteins: a bacterial plasmid designed for protein expression, and a baculovirus system. Which system would be her best choice to successfully clone and express (a) the gene encoding the E. coli RecA protein, and (b) the gene encoding a mammalian DNA polymerase?
Question 7.14
In the protocol for Western blots, an investigator uses two antibodies. The first binds specifically to the protein of interest. The second is labeled for easy visualization and binds to the first antibody. In principle, molecular biologists could simply label the first antibody and skip one step. Why do they use two successive antibodies?
Question 7.15
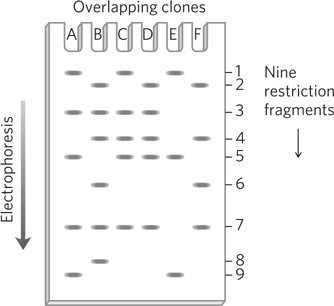
A group of overlapping clones, designated A through F, are isolated from one region of a chromosome. Each of the clones is separately cleaved by a restriction enzyme, and the pieces are resolved by agarose gel electrophoresis, with the results shown below. Nine different restriction fragments can be produced from this chromosomal region, with a subset appearing in each clone. Using this information, deduce the order of the restriction fragments in the chromosome.
Question 7.16
You are a researcher who has just discovered a new protein in a fungus. To help determine the protein’s function, you want to identify the other proteins in the fungal cell with which your protein interacts. How do you design a yeast two-
Question 7.17
Figure 7-30 shows the first steps in the process of making a DNA microarray by photolithography. Describe the remaining steps for obtaining the desired sequences (a different four-
Question 7.18
Mammalian genomes contain some short sequences repeated over and over again. Thousands of these sequences are sometimes repeated in tandem (e.g., the sequence TTAGGG repeated thousands of times in succession). When these tandem repeat regions exceed a few hundred base pairs, it becomes difficult to define them adequately using next-
256
Question 7.19
In the CRISPR/Cas9 nuclease system, what is the role of the sgRNA?