Chapter Introduction
14: Site-Specific Recombination and Transposition
MOMENT OF DISCOVERY

Wei Yang
I arrived at Yale University as a postdoctoral fellow in the laboratory of Dr. Tom Steitz 10 years after he and Nigel Grindley solved the crystal structure of the catalytic domain of γδ resolvase, a site-specific recombinase. My goal was to crystallize a protein-DNA complex to aid in understanding the recombination mechanism. The DNA recombination site res is 114 base pairs long. However, two res sites must be in supercoiled DNA for recombination to take place, making the complex very difficult to crystallize.
In the early 1990s, we had to use a “divide and conquer” strategy. We started by co-crystallizing DNA containing only one res cleavage site with a dimeric γδ resolvase. It was terribly exciting for me to find the initial crystals while peering into a microscope, but the experienced postdocs and students in Steitz’s lab just smiled and politely wished me luck. I understood their reaction only after finding that many protein-DNA co-crystals don’t diffract x rays well or at all.
After overcoming the diffraction problem, the phase problem, and the asymmetry of the γδ resolvase dimer, the moment of visualizing the electron density of the γδ resolvase–DNA complex on a computer screen for the first time was sheer joy. At that moment I was the only one in the whole world who knew how γδ resolvase bound (and bent!) its recognition site. However, obtaining the crystal structure of a true recombination intermediate took another 10 years.
Today, more than 15 years after I solved the γδ resolvase–DNA complex structure, the mechanism of how two DNA duplexes exchange partners remains a hypothesis. To fully understand a biological process, it often takes generations of scientists, with each generation making additional steps forward. I was pleased to contribute my step and eagerly look forward to following the next chapters in this story.
—Wei Yang, on researching the structure and molecular mechanisms of γδ resolvase
Life might have made its first appearance on this planet in the form of a self-replicating polymer made of RNA, or something similar. The existence of this successfully self-replicating RNA would immediately have given rise to a biosphere with two possible survival strategies. First, the polymer itself could continue to use the strategy of self-replication, drawing on resources in the environment to increase its own population. Second, in a strategy that introduced what were probably the first parasites, a smaller RNA—a primitive RNA transposable element—could insert itself into a vulnerable site in a self-replicating RNA. With this strategy, the transposable element would reproduce passively as long as the function of the self-replicator was unaffected by the addition to its mass. Thus, transposable elements may be among the most ancient of genetic entities. Always present and often deceptively simple in structure, transposons long ago perfected the art of mostly benign coexistence with their hosts. Some bear a close evolutionary relationship to viruses. The additional RNA contributed by the parasitic transposable elements was not always inconsequential; transposable elements have had a profound effect on living systems throughout evolutionary time. For example, they played a key role in the development of the vertebrate immune system, as we will see.
Perhaps most important, the fundamental chemistry employed by these elements is seen today in processes ranging from the hydrolytic cleavage of nucleic acids (Chapter 6) to DNA polymerization (Chapter 11), topoisomerization (Chapter 9), RNA transcription (Chapter 15), and intron splicing (Chapter 16). The basic reaction underlying all of these processes is a phosphoryl transfer involving a phosphodiester bond (Figure 14-1). Many of these processes represent a specialized form of transesterification. It is the phosphoryl transfer reactions that ultimately make every nucleic acid a dynamic purveyor of the genetic information on which all life depends. We find that basic chemistry not only in transposition but in every process described in this chapter, including site-specific recombination and the integration of retroviruses (including HIV) into genomic DNA.
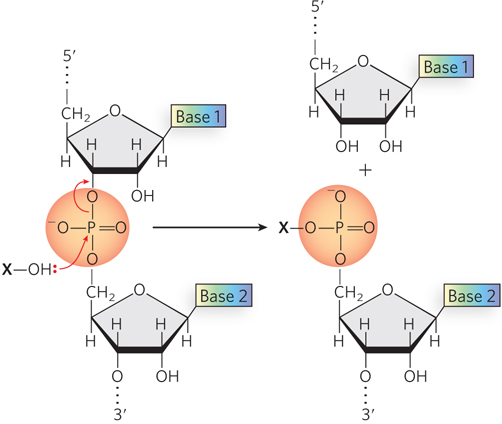
Figure 14-1: Phosphoryl transfer reactions involving phosphodiester bonds. In three common variants of this reaction described in this chapter, the nucleophilic hydroxyl group (X–OH) is (1) water, leading to hydrophilic cleavage; (2) the hydroxyl group of a Ser or Tyr residue in the active site of an enzyme, leading to the formation of a covalent phosphodiester linkage to that enzyme at the expense of the existing phosphodiester bond between nucleotides; or (3) the 3′-hydroxyl group of another nucleotide, leading to formation of a new phosphodiester bond at the expense of the existing one. The second and third processes are generally isoenergetic, or nearly so. The closely related phosphoryl transfer reactions leading to formation of phosphodiester bonds in the active sites of DNA and RNA polymerases are illustrated in Chapters 11 and 15, respectively.
Site-specific recombination and transposition are specialized types of recombination that share two key properties. First, both generally involve bringing together DNA sites without extensive homology. In that sense they complement the homologous genetic recombination discussed in Chapter 13, expanding the repertoire of DNA and RNA transactions available to every cell. Second, the key enzymes in both processes bear a notable evolutionary relationship to DNA topoisomerases. Site-specific recombinases and transposases inhabit an enzymatic world of thermodynamic equanimity. Like topoisomerases, they promote phosphoryl group transfers that are typically isoenergetic (i.e., ΔG′° = 0), or nearly so.
Site-specific recombination is a precise and predictable process in which DNA is rearranged between two specific sequences. This can result in the insertion, deletion, or inversion—depending on the arrangement of the recombination sites—of a particular DNA segment. In a sense, recombinases, the enzymes that carry out the site-specific recombination reactions, are restriction endonucleases and DNA ligases combined into a single efficient package. However, unlike DNA ligation, the reactions do not require ATP or a similar cofactor; the phosphoryl transfer reactions underlying site-specific recombination are nearly isoenergetic. The reactions are often tied to genomic replication, but they have been recruited for other purposes as well. They typically provide an elegant biochemical solution to awkward topological problems that occur in plasmids and chromosomes. Site-specific recombinases are increasingly well understood, and biotechnologists love them for the precise DNA rearrangements they catalyze.

Barbara McClintock, 1902–1992
Transposable elements, also called transposons, are much more than the hypothetical denizens of an early RNA world. They exist today, tucked away in the genomes of essentially all organisms. They are genomic freeloaders, nucleic acid sequences that insert themselves into the genome of another organism and are replicated passively every time a cell divides. They are almost stunningly common: as noted in Chapter 8, the genomes of several million transposons or transposon remnants make up nearly half of the DNA in the human genome. Through the activity of enzymes called transposases, these genetic elements can move from one location on a chromosome to another, often with no apparent selectivity with respect to the surrounding DNA sequence. This process is known as transposition. The ability of transposons to move within a genome, defying Mendelian genetics, startled and then intrigued geneticists when Barbara McClintock discovered these elements in maize plants in the 1940s. Careful work by McClintock, and other researchers’ eventual discovery of closely related genetic elements in other organisms, gradually overcame the initial skepticism of colleagues. Since then, studies of transposons have placed them among the most important tools of biotechnology, yielding invaluable genetic techniques for constructing transgenic model organisms and for exploring gene function. As noted above, transposons insert themselves into genomic DNA through nearly isoenergetic phosphoryl transfer reactions like that shown in Figure 14-1.
In this chapter, we discuss site-specific recombination and transposition in succession. We begin with site-specific recombination because it is more predictable and allows us to layer biochemistry and biology onto the chemistry of Figure 14-1 in a more intuitive progression.


