14.1 MECHANISMS OF SITE-SPECIFIC RECOMBINATION
Recombination between specific sequences can result in insertion, deletion, or inversion of the DNA sequence between those sites. Recombination reactions of this type occur in virtually every cell, filling specialized roles that vary greatly from one species to another but sharing a common mechanism. Each site-
The DNA rearrangements promoted by site-
Precise DNA Rearrangements Are Promoted by Site-Specific Recombinases
The recombination sites recognized by site-
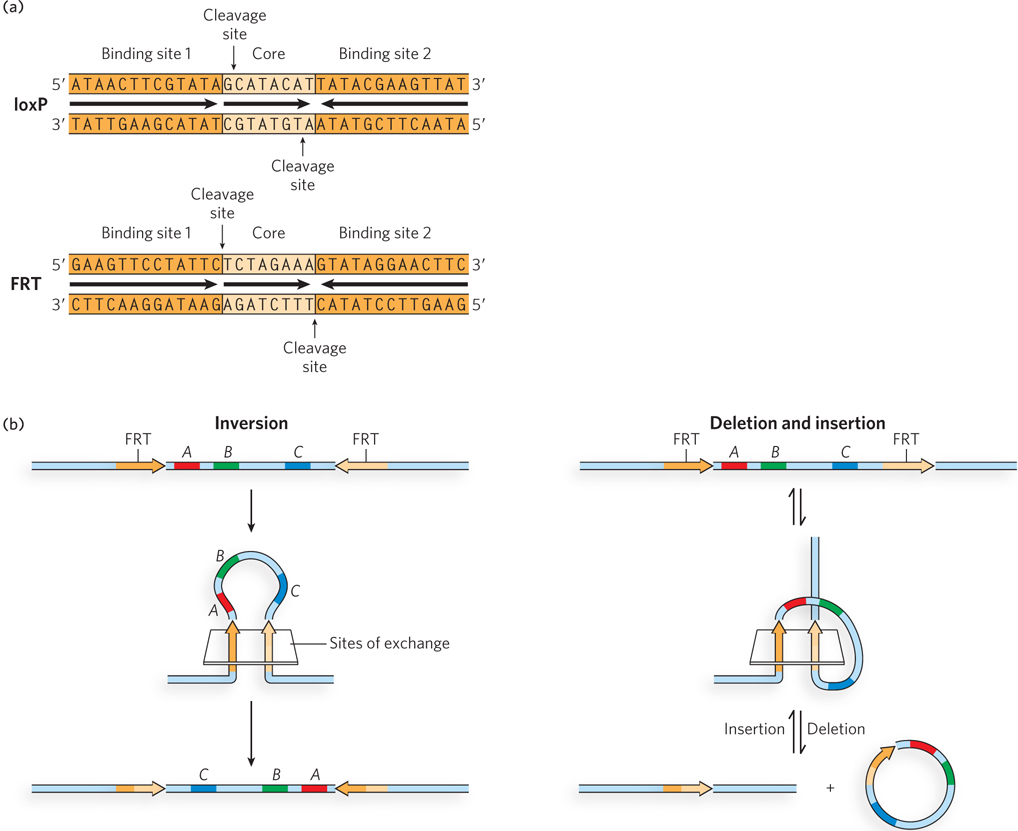
The asymmetric core sequence in a recombination site gives each site an orientation within the surrounding DNA. The overall outcome of a site-
Site-
The site-
488
489
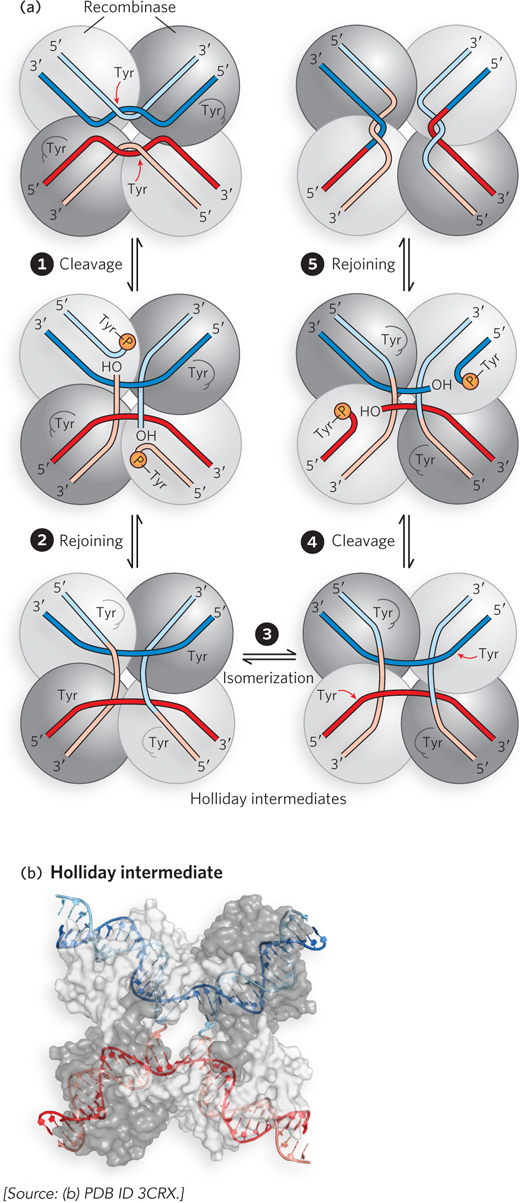
In systems that use an active-
Many mechanistic details of these reactions have become clear with the structural elucidation of recombinases caught at different steps of the process. The four recombinase subunits and the four DNA arms in the synaptic complex take up a square planar arrangement (Figure 14-3b). As shown by the crystal structure, the tyrosine-
The overall process is closely related to the reaction mechanism promoted by topoisomerases (see Figure 9-19). For both topoisomerases and site-
490
Site-Specific Recombination Complements Replication
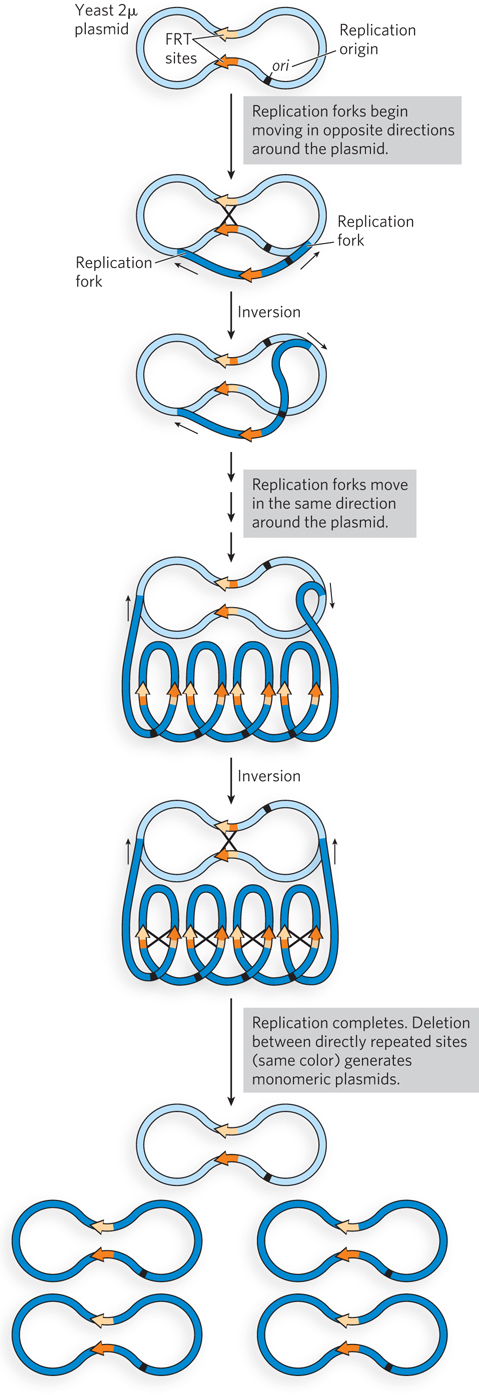
Replication of the circular chromosomes of viruses, plasmids, and many bacteria poses a unique set of challenges. As noted in Chapter 13, recombinational DNA repair of stalled replication forks can give rise to contiguous dimeric chromosomes (see Figure 13-14). A specialized site-
A common plasmid in Saccharomyces cerevisiae, the 2μ (2 micron) plasmid, has a site-
491
Site-Specific Recombination Can Be a Stage in a Viral Infection Cycle
Bacteriophages such as P1 and λ have played important roles in the development of molecular biology and biotechnology (see the How We Know section at the end of this chapter). When it enters a cell, the DNA of these phages has two potential fates (Figure 14-5). A lysogenic pathway involves incorporation of the phage genome as part of the host genome, either by integration into the host chromosome or as an autonomously replicating plasmid. In either case, phage genes are largely repressed, and the phage DNA is replicated passively by host enzymes. Lysogenized bacteriophage genomes are referred to as prophages, and the parasitic infection is benign as long as the phage remains in this state. In the alternative, lytic pathway, the bacteriophage DNA is replicated and packaged into new phage heads, and the host cell is destroyed by lysis to disperse the progeny. The specific mechanisms used in the P1 life cycle feature site-
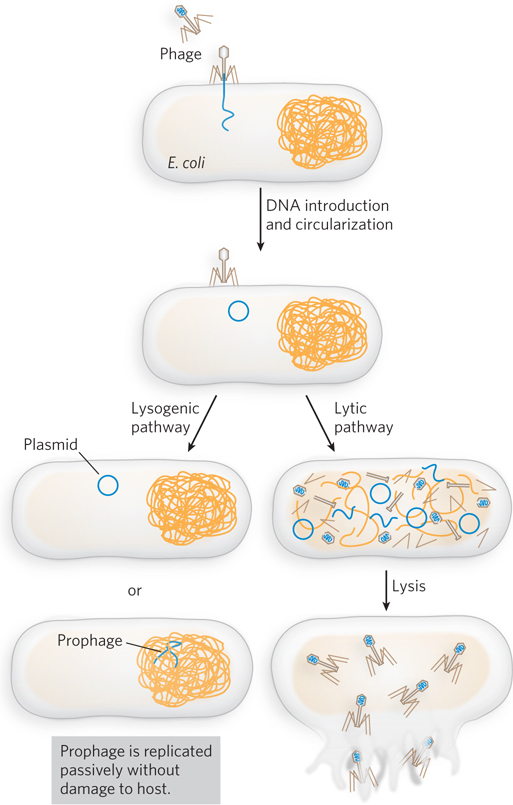
P1 enters a bacterial cell as a linear DNA molecule containing multiple contiguous copies of the 90 kbp genome. In the host, the DNA is rapidly circularized to produce multiple genome-
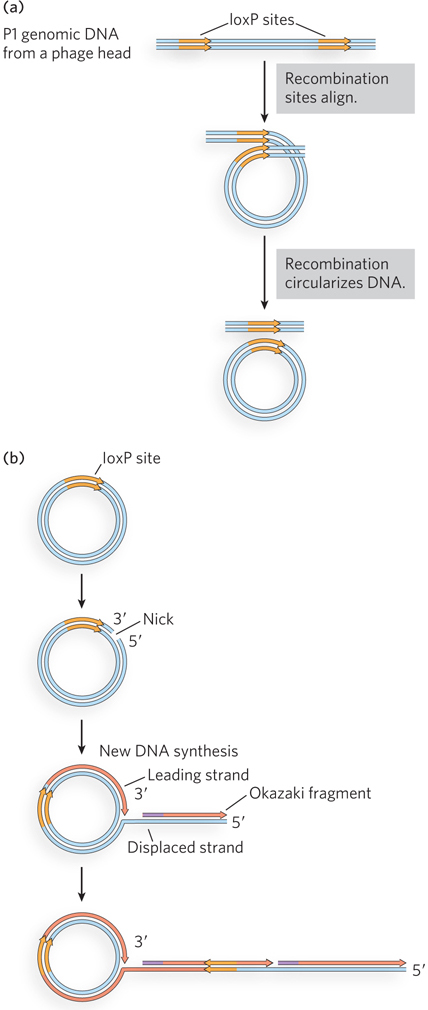
Site-Specific Recombination Systems Are Used in Biotechnology
As noted earlier, the Flp recombination system of the yeast 2μ plasmid and the Cre-
492
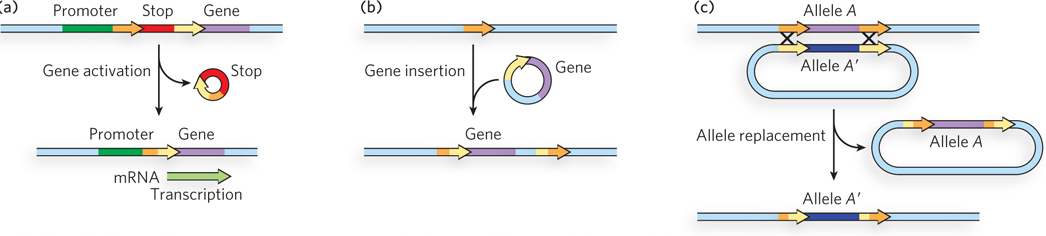
The Cre and Flp systems will function when engineered into the cells of any organism and thus are highly useful in a wide range of biotechnological applications. A few such applications are shown in Figure 14-7. If the requisite lox or FRT sites are engineered into plasmids or chromosomes in the proper locations, these systems can be (and have been) used to activate a particular gene, insert a new gene into a cell at a chosen location, replace one gene with another gene or an altered version of the same gene, delete a gene, or alter the linear structure of an entire chromosome. The sequence specificity of the recombinases allows all of these transactions to be promoted with extraordinary precision. Even more elaborate manipulations are possible. For example, if you tied expression of the recombinase to a promoter expressed only in a particular tissue, you could limit the recombination event to that tissue. Deleting a gene in a certain tissue at a specified time can be a powerful tool for exploring the function of that gene. A vivid example of the application of site-
Gene Expression Can Be Regulated by Site-Specific Recombination
The biological uses of site-
The switch is accomplished by periodic inversion of a segment of DNA containing the promoter for a flagellin gene. The inversion is a site-
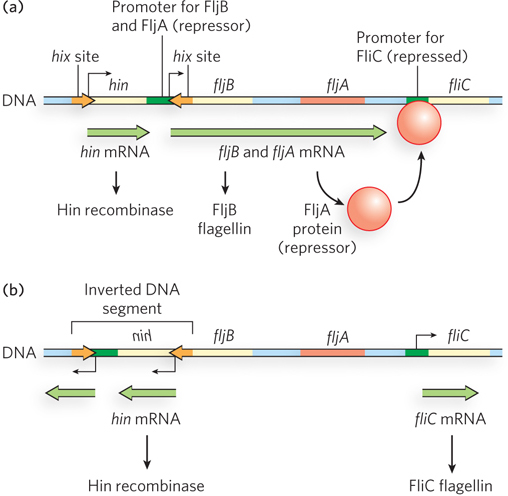
493
The Salmonella system is by no means unique. Similar regulatory systems occur in other bacteria and in some bacteriophages. Gene regulation by DNA rearrangements that move genes and/or promoters is particularly common in pathogens that benefit by changing their surface proteins, thereby staying ahead of host immune systems.
Hin belongs to a family of recombinases that are distinct from the Flp and Cre enzymes: the serine-
The Hin recombinase catalyzes recombination only when its hix recombination sites are on the same supercoiled DNA molecule and are inverted relative to each other. This specificity is accomplished through the formation of an elaborate structure involving both Hin and the Fis proteins, which acts as a topological filter (Figure 14-9). The structure cannot form unless the DNA is supercoiled and the sites are properly oriented. The basic site-
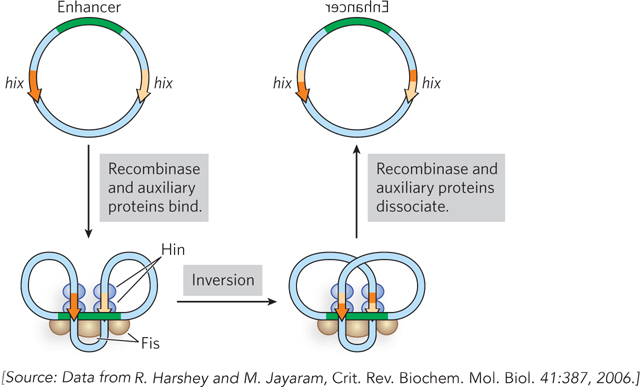
494
HIGHLIGHT 14-1 TECHNOLOGY: Using Site-Specific Recombination to Trace Neurons
Site-
The use of site-
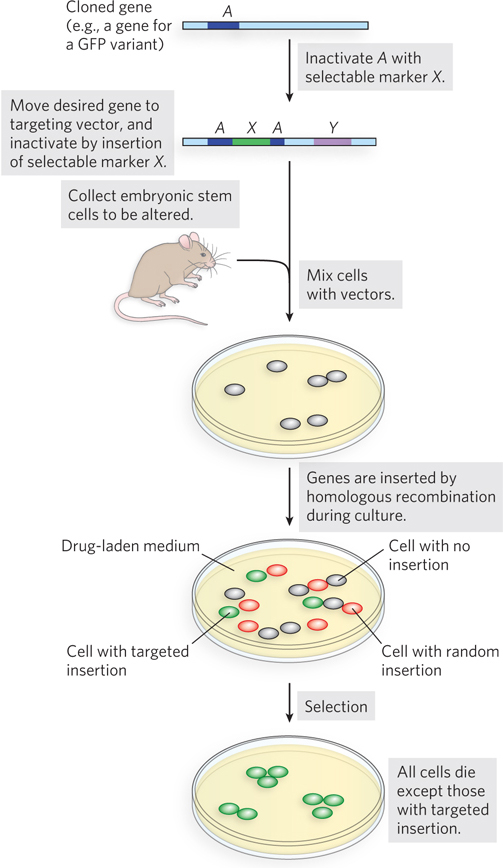
Mario Capecchi, Oliver Smithies, and Martin Evans developed powerful procedures to generate transgenic mice with designed genomic alterations (Figure 1; see Chapter 7 for discussion of such procedures). DNA containing a gene alteration is introduced into mouse embryonic stem cells. The DNA is introduced in the form of an engineered plasmid DNA that sandwiches the desired alteration between a set of selectable markers that will allow identification of the rare cells with the desired alteration inserted at the correct location. The altered stem cells (from brown mice) are introduced into a very early embryonic stage (blastocyst) of black mice, and the embryos are implanted in a surrogate mother. The altered brown-
The brainbow method makes use of green fluorescent protein (GFP), the procedures for generating transgenic mice, and site-
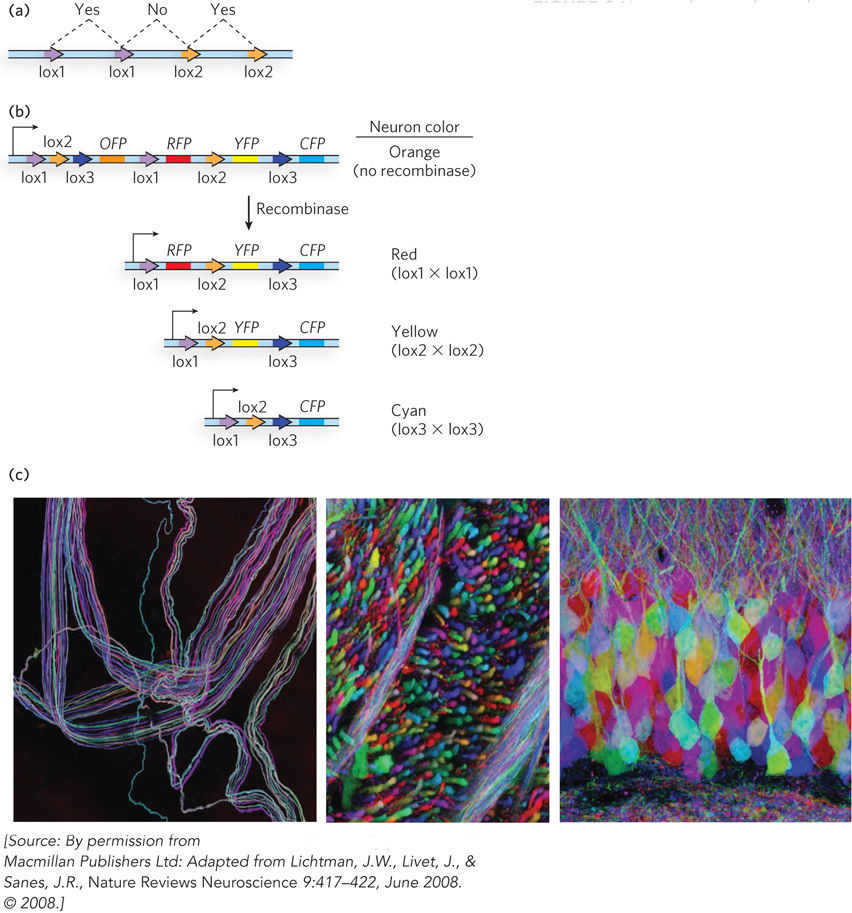
495
The engineered mice with GFP cassettes are homozygous for these cassettes, and they pass them on to all their progeny. Separately, a second population of homozygous transgenic mice is engineered to express the Cre recombinase, again from a promoter directing gene expression transiently and only in developing neurons. When a mouse with the GFP cassettes is mated to a mouse expressing the Cre recombinase, all progeny are heterozygous for both the cassette and the recombinase genes. As these mouse embryos develop, the Cre recombinase is expressed early in the development of each neuron. Recombination events occur in some or all of the cassettes in a given neuron, leading to the expression of a particular set of GFP variants. Mixing several different GFP variants in a cell increases the number of potential colors. The outcome is unpredictable for each cell. However, only one set of recombination reactions occurs in each cell, and the end result imparts a distinctive color that is expressed for the life of that neuron. Neighboring developing neurons go through the same recombination processes, but the number of possible outcomes is large and neighboring cells rarely acquire the same color. The result is a rainbowlike array of fluorescent colors in the neural network—
496
SECTION 14.1 SUMMARY
Site-
specific recombination entails the precise cleavage and rejoining of DNA ends at specific and reproducible sites in the DNA. There are two classes of site-
specific recombinases, defined by the key nucleophilic amino acid residue at their active sites: tyrosine or serine. Site-
specific recombination can be coupled to replication, or resolution of chromosomal dimers to monomers before cell division, or amplification of plasmid copy number. The mechanism of site-
specific recombination can be used to facilitate DNA transactions critical to a viral life cycle. Biotechnologists have adapted site-
specific recombination systems to manipulate DNA segments ranging from plasmids to genomes. Some organisms use site-
specific recombination to regulate gene expression, as in phase variation in Salmonella. Site-
specific recombination can be rendered specific for one particular reaction outcome (integration, deletion, or inversion) by coupling the reaction to the formation of a larger, structured complex in which only one reaction outcome is possible.