DATA ANALYSIS PROBLEMS
Chapeville, F., F. Lipmann, G. Von Ehrenstein, B. Weisblum, W.J. Ray Jr., and S. Benzer. 1962. On the role of soluble ribonucleic acid in coding for amino acids. Proc. Natl. Acad. Sci. USA 48:1086–
Question 18.14
The experiments demonstrating that ribosomes check the anticodon of the tRNA, but not the amino acid attached to it, were carried out well before the entire genetic code was defined. The research team, led by Seymour Benzer (see the How We Know Section in this chapter), chose tRNACys for their demonstration, for several reasons—
Which amino acids are incorporated into a polypeptide specified by a random sequence of U and G?
Why was tRNACys a good choice for the experiment?
Benzer and colleagues showed that incorporation of radioactively labeled amino acids into polypeptide was linearly related to the amount of labeled aminoacyl-
tRNA added to the experiment. They treated one preparation of [14C]Cys- tRNACys with Raney nickel; about 60% of the charged tRNA was reduced to [14C]Ala- tRNACys. They then plotted incorporation of radioactivity, over time, in preparations with [14C]Cys- tRNACys (Figure 1) and preparations with [14C]Ala- tRNACys (Figure 2), measured in counts per minute (cpm) in the trichloroacetic acid– precipitated (TCA- ppt) preparations. Given the amount of labeled aminoacyl-
tRNA added (indicated on the vertical axis of the graphs) and the amount incorporated into polypeptide, does this result support the contention that Ala- tRNACys is incorporated at codons specifying Cys? Why or why not? The researchers next treated a preparation of [14C]Phe-
tRNAPhe with Raney nickel, then compared the incorporation of [14C]Phe into polypeptide in response to a poly(U) RNA before and after Raney nickel treatment. No effect of the Raney nickel treatment was seen. 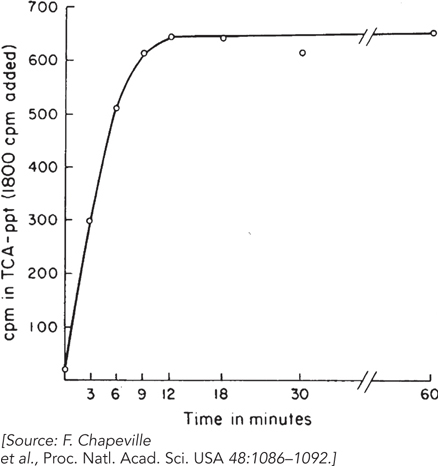 FIGURE 1 [14C]Cys-
FIGURE 1 [14C]Cys-tRNACys incorporation. 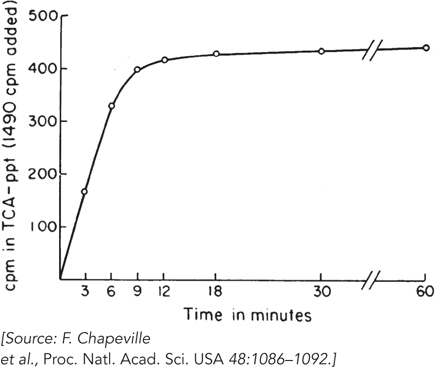 FIGURE 2 [14C]Ala-
FIGURE 2 [14C]Ala-tRNACys incorporation. Why was this experiment carried out?
The polypeptides synthesized using poly(UG) and Ala-
tRNACys were digested to single amino acids, and the products were analyzed to directly demonstrate that about 60% of the labeled residues were Ala.
Noller, H.F., V. Hoffarth, and L. Zimniak. 1992. Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256:1416–
Question 18.15
Experiments strongly suggesting that the peptidyl transferase activity of ribosomes is due to rRNA, not r-
663
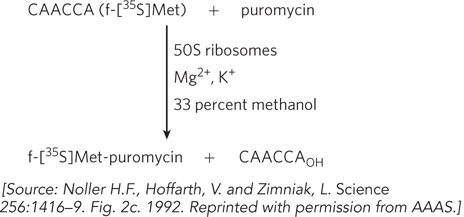
What do the substrates required for this in vitro reaction suggest about the binding properties of the 50S ribosomal subunit?
What is special about the hexanucleotide sequence attached to the labeled fMet residue?
In which ribosomal site must the fMet-
oligonucleotide bind, and in which site must the puromycin bind? What advantages does this reaction have for this particular study, relative to protein synthesis with complete ribosomes and mRNA and charged tRNAs?
Using this assay, the researchers demonstrated that the 50S subunit or the intact 70S ribosome from E. coli would catalyze the reaction even after extraction with sodium dodecyl sulfate (a detergent that denatures proteins) and extensive treatment with proteinase K (which degrades virtually all proteins). Extraction with phenol (a treatment that separates protein from nucleic acid), however, led to a loss in activity. Subsequently, the investigators found that 50S subunits derived from the thermophilic bacterium Thermus aquaticus did not lose activity when extracted with phenol.
Why did the researchers explore the 50S subunits from T. aquaticus?
Noller and colleagues went on to test the sensitivity of the reaction to chloramphenicol, carbomycin (another ribosome-
binding inhibitor, with a mechanism similar to chloramphenicol), and a general ribonuclease called RNase T1, with or without (+ or −) SDS, proteinase K (PK), or phenol treatment. The results are shown in Figure 2, an autoradiograph of the product after high- voltage paper electrophoresis. 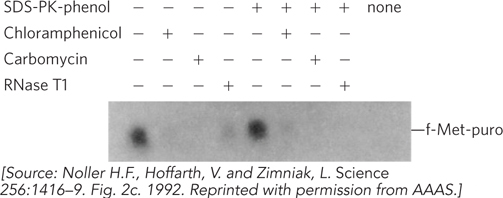 FIGURE 2
FIGURE 2What conclusions can you derive from the results of this experiment?
The conclusion that ribosomes are ribozymes did not achieve general acceptance until elucidation of the three-
dimensional structure of a bacterial 50S ribosomal subunit. Suggest a reason for the delay in general acceptance.