20.3 CONTROL OF GENE EXPRESSION IN BACTERIOPHAGES
Our discussion of gene regulatory pathways up to this point has focused on bacteria. Historically, however, much of the early research on gene regulation was done on viruses—
We focus here on the bacteriophage λ (λ phage) system, one of the best-
716
Phage Propagation Can Take One of Two Forms
Most organisms are susceptible to infection by viruses, which usurp the host cell machinery to produce more viral particles. One well-
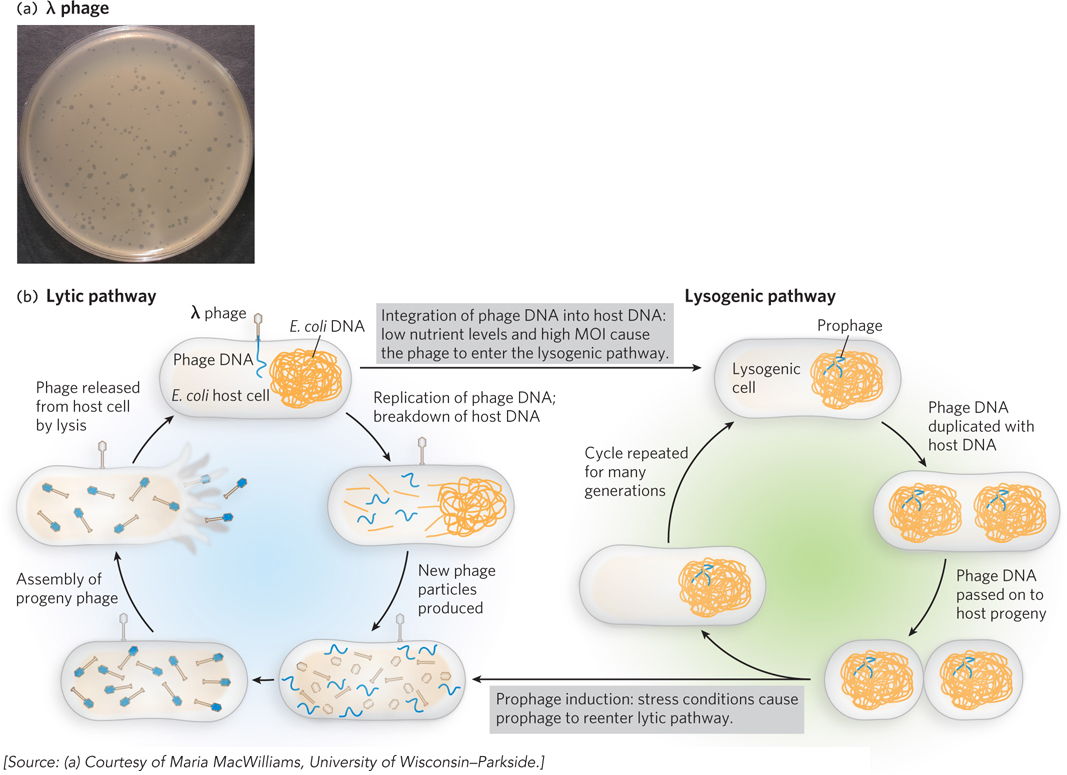
717
Many phages are capable of this kind of genetic switch, but the underlying mechanisms are best understood for λ phage, a virus of E. coli that consists of a double-
Two regulatory proteins, the λ repressor (also called cI) and Cro, govern the growth pathway of the virus. When the λ repressor protein is predominant, λ phage enters the lysogenic pathway; its DNA integrates into the chromosome, and only the λ repressor itself is expressed. When Cro predominates, however, λ phage enters the lytic pathway; most of the λ genes are expressed, and viral replication and packaging ensue. Eventually, the host cell is broken open by cell lysis, a process that releases the progeny phages.
Whether the phage enters lytic or lysogenic growth is determined early in a λ phage infection and depends largely on two other λ proteins: cII and cIII. The cII protein is a transcription activator that enhances transcription of the λ repressor gene and hence stimulates production of the repressor protein (cI). When cII is abundant and active, infection proceeds through the lysogenic pathway. But because cII is susceptible to degradation by bacterial proteases, it is often destroyed before it can trigger substantial production of the λ repressor. This tends to occur under nutrient-
The cIII protein stabilizes cII, probably by acting as a decoy, or alternative substrate, for protease molecules that would otherwise degrade cII. In this way, elevated cIII levels can trigger the switch to lysogen formation by enhancing cII concentrations, which increases production of the λ repressor.
Usually, λ phage infection leads to lytic growth. In addition to nutrient levels, the multiplicity of infection (MOI) also affects this growth pathway. This is because infections typically occur at a low MOI, when there are many more host cells than viral particles. In this situation, it is advantageous for viruses to grow lytically so that more progeny can be made and released to infect the available host cells. However, once there is an abundance of viral particles relative to host cells, multiple viruses begin to infect each host. In this circumstance, the high MOI leads to more frequent lysogen formation. The virus propagates silently within the chromosome and awaits future opportunities to enter the lytic pathway when host cells are again abundant. The mechanisms underlying this fascinating genetic switch have been elucidated over many years, using numerous genetic and biochemical methods.
Differential Activation of Promoters Regulates λ Phage Infection
Most of the ∼50 genes of the λ phage genome are required for viral replication and packaging; a relatively small region of the genome is critical for the gene regulation necessary to induce lytic versus lysogenic growth (Figure 20-21). On initial infection of a host cell by λ DNA, viral transcription begins at two opposing promoters, PL (leftward promoter) and PR (rightward promoter), to produce the “immediate early” transcripts. This leads to the production of two proteins, N and Cro, which begin to increase in concentration. Two additional promoters, PRM (promoter for repressor maintenance) and PRE (promoter for repressor establishment), drive transcription of the cI gene, which encodes the λ repressor. However, unlike the strongly constitutive PL and PR, PRM and PRE are weak promoters and require activators to recruit RNA polymerase. Thus, at first, the cI gene is not transcribed and no λ repressor is produced.

718
Overlapping the PL, PRM, and PR promoters are two operator sites, OL and OR, each of which contains three binding sites for the λ repressor and Cro. Like the Lac repressor, both the λ repressor and Cro form homodimers in which two DNA-
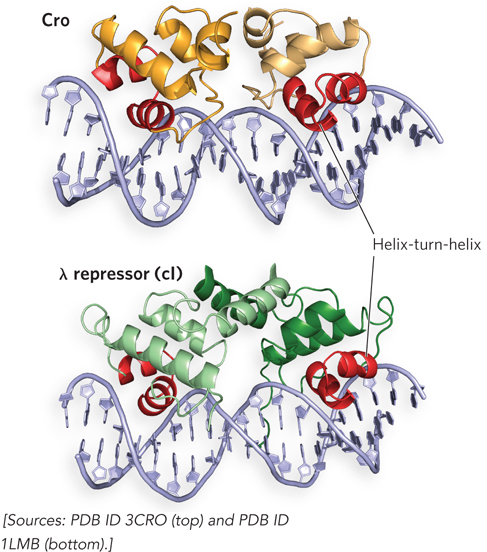
During initial viral infection, once appreciable levels of Cro are expressed, this protein binds the OR3 site; because OR3 overlaps PRM, Cro blocks the access of RNA polymerase to PRM, and the cI gene is not transcribed. Cro does not bind as well to OR1 and OR2, and thus these operator sites are not occupied by Cro. If this situation continues, lytic growth ensues (Figure 20-23a).
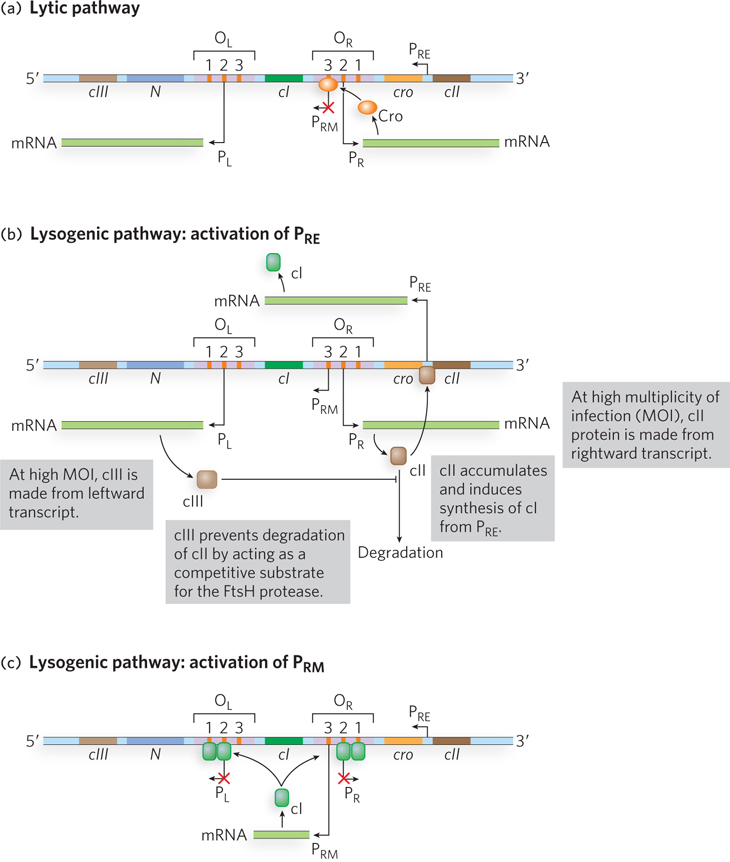
Lysogenic growth requires the activity of PRE (Figure 20-23b). Initial transcription of the cI gene requires activation of PRE by cII, another gene product expressed early in the infection process. The cII protein can activate the weak PRE promoter by binding to a site upstream from the transcription start site (the −35 region; see Chapter 15) and recruiting RNA polymerase to PRE. However, cII is a substrate of the host cell protease FtsH, which cleaves and inactivates the activator. At a low MOI, in which one viral particle at most has infected any one cell, the concentration of cII does not reach a level sufficient to activate PRE. But under conditions of high MOI, in which multiple viral particles have infected the cell, more cII is produced because of the multiple copies of the cII gene present in the cell. Furthermore, the phage protein cIII (also expressed early in infection) helps stabilize cII by competing as a substrate for FtsH. In this situation, FtsH cannot keep up with cII production, and the increased levels of cII lead to activation of PRE.
As a result of the activation of PRE, the λ repressor protein is produced. The repressor activates PRM by binding to OR1 and OR2, leading to stable expression of the repressor (Figure 20-23c). The PRM promoter is necessary to maintain ongoing production of the λ repressor because, when the repressor binds operator sites OR1 and OR2, PR is silenced and production of cII stops, leading to loss of the activator (cII) for PRE. Under these conditions, lysogenic growth is favored.
Note that if Cro were to bind OR3 before cI (the repressor) bound OR1 and OR2, the phage would have trouble establishing lysogeny. This interference by Cro probably never occurs, however, because when cII is highly active, cI production is sufficient to shut off the cro gene before enough Cro is made to turn off PRM.
The λ Repressor Functions as Both an Activator and a Repressor
The λ repressor is capable of complex regulation, in part because it is a two-
Once the lytic-
719
More Regulation Levels Are Invoked during the λ Phage Life Cycle
Switching between the activation and repression of promoters enables λ phage to alternate between lytic and lysogenic growth, as dictated by environmental conditions. But additional levels of regulation further control the expression of genes required to establish and maintain early steps in the infection cycle. Two λ proteins, the N and Q proteins, are known as antiterminators because they prevent RNA polymerase from prematurely stopping transcription of the genes they regulate. N protein binds to specific regions of the viral transcript called nut (N utilization) sites (Figure 20-24). The resulting RNA-
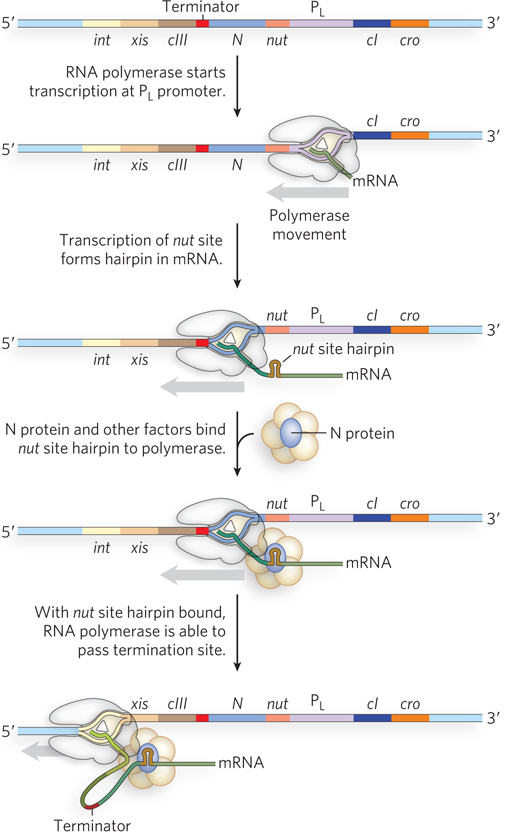
720
One target of the N protein antitermination mechanism is the viral gene encoding the Q antiterminator protein. Unlike N, Q binds DNA sequences between the −10 and −35 regions of PR′, the promoter for genes that are active late in the infection process. In the absence of Q, RNA polymerase pauses shortly after initiation of the transcript, then falls off the template when encountering a terminator hairpin structure, 200 bp further on. When Q is present, it first binds in the promoter region, then transfers to the polymerase when the enzyme pauses after initiation. With Q bound, the polymerase can transcribe through the terminator hairpin, thereby producing full-
To establish a lysogenic infection, the virus must produce an integrase, an enzyme responsible for integrating phage DNA into the host cell chromosome (see Chapter 14). The int gene, which encodes the integrase, is transcribed from two promoters, PI and PL. The cII protein activates PI, in addition to PRE (as described earlier); thus, when cII is abundant and the lysogenic state is favored, both repressor and integrase are expressed. Although the int gene is also transcribed from PL, the two different transcripts have inherently different stabilities due to different RNA structures at their 3′ ends. The PI-derived transcript ends at a terminator hairpin structure downstream from the integrase-
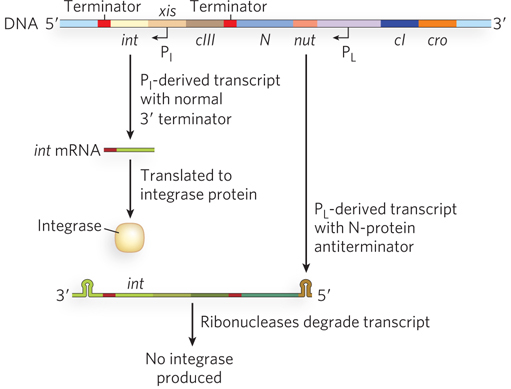
721
In a twist on this regulatory mechanism, the λ DNA integrates into the host chromosome such that a small portion of the phage DNA is removed—
SECTION 20.3 SUMMARY
Bacteriophage gene activities can be controlled by regulatory networks that integrate multiple signals into a common gene regulatory response.
Infection of a bacterial cell by λ phage can lead to either (1) a lytic pathway, in which host cells are lysed as new viral particles are assembled and released, or (2) a lysogenic pathway, in which the viral DNA integrates into the host chromosome and is propagated through chromosome replication and cell division, without immediate production and release of new viruses.
In λ phage, two regulators, Cro and the λ repressor, control whether the phage propagates through the lytic pathway (when Cro dominates) or the lysogenic pathway (when the λ repressor dominates). Both repressors bind to similar phage DNA sequences, but with different binding affinities. Cro blocks λ repressor synthesis while allowing expression of other genes needed for lytic growth; the λ repressor blocks transcription of all phage genes except its own.
In addition to Cro, the N protein, expressed early in the infection process, favors λ gene expression by binding nut sites in the viral transcripts. The resulting RNA-
protein complexes favor transcription by assembling with host proteins that aid binding of N protein to RNA polymerase, enhancing the enzyme’s ability to bypass terminator structures downstream from the N and Cro genes. N protein–
mediated antitermination favors production of the Q protein, another antiterminator that binds DNA sites near the promoter of genes expressed later in the infection cycle. Binding of the Q protein allows it to transfer to paused RNA polymerase molecules and enhance their ability to traverse terminator structures in the regulated genes. To establish a lysogenic infection, the virus produces an integrase that integrates the phage DNA into the host chromosome. Integrase is also required to excise the prophage from the host genome.
UNANSWERED QUESTIONS
The study of gene regulation continues to expand as new regulatory mechanisms are uncovered. RNA has emerged as a major player in controlling levels of protein production, and many fascinating aspects of its involvement remain to be deciphered.
How widespread are RNA-
based gene regulatory mechanisms? Early work on the regulation of gene expression focused on proteins that have the sole role of controlling when and how much of a particular protein is synthesized. More recent research has revealed an increasing number of instances in which RNA plays this regulatory role. In addition to the use of riboswitches, bacteria produce many small noncoding RNAs that are important for modulating gene expression. Understanding how these work and how their mechanisms relate to those responsible for regulating gene expression in plants and animals (see Chapter 22) is a fascinating area of research. How does a bacteriophage compete for a host cell’s gene expression machinery? Studies of λ phage have provided an exciting introduction to the world of phage–
host cell competition, but there are many different mechanisms by which viruses might take over a host cell’s gene expression machinery and thus regulate the propagation of new viral particles. Some researchers estimate that Earth is home to more phage particles than cells! Phages thus provide an enormous pool of genes that are readily exchanged and introduced into new hosts, driving the evolution of new traits and viral defense mechanisms. A broader understanding of gene regulatory pathways in phages will offer new insights into bacterial gene regulation, and perhaps into the relationships between viral propagation and gene transfer. How are gene regulatory networks integrated? Much of the research on gene regulation in bacteria has focused on individual genes or operons, giving the impression that just one or a few changes occur in response to signaling molecules. Studies using DNA microarrays and high-
throughput sequencing, however, show that changes in gene expression in response to stresses or altered nutrient levels occur in hundreds of different genes. How these changes are coordinated and how different gene regulatory pathways integrate multiple signals at once are the subjects of active research. Investigators are using traditional genetic and biochemical methods, as well as approaches including high- throughput RNA sequencing (RNA- Seq) and bioinformatics. This area of research is referred to as systems biology, to indicate that gene expression operates not in isolation but as part of a system, as defined by the cell or organism.
722
TRAPped RNA Inhibits Expression of Tryptophan Biosynthetic Genes in Bacillus subtilis
Babitzke, P., and P. Gollnick. 2001. Posttranscription initiation control of tryptophan metabolism in Bacillus subtilis by the trp RNA-
The TRAP system of tryptophan regulation in B. subtilis beautifully illustrates how the mechanistic details of multilevel gene regulation were eventually worked out using a combination of experimental methods. Similar to the process in other bacteria, the trp genes of B. subtilis, contained in the trpEDCFBA operon (each letter after trp standing for a gene in the operon), are transcribed only when tryptophan is in short supply in the cell. Genetic experiments revealed that transcription of the trpEDCFBA operon requires a regulatory protein called TRAP (trp RNA–
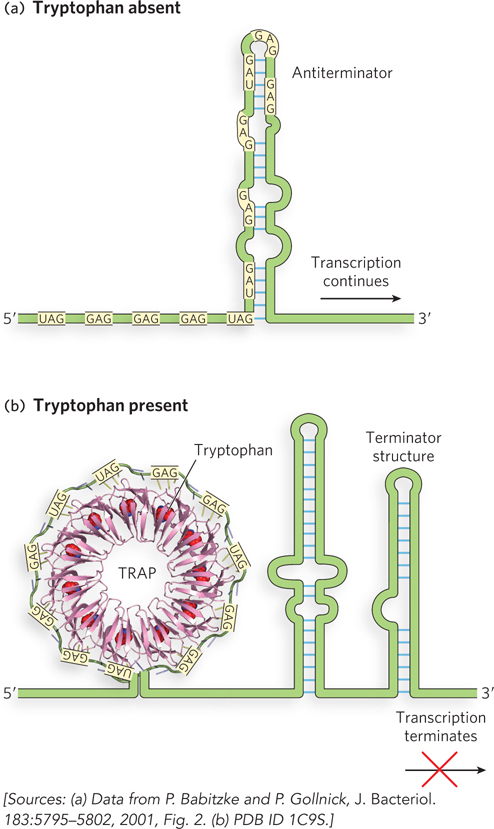
How does TRAP respond to l-tryptophan? TRAP is composed of 11 subunits, each of 6 to 8 kDa. TRAP binds to 11 triplet repeats, primarily GAG and UAG, in the leader sequence that are separated from each other by two or three nonconserved nucleotides. The crystal structure of TRAP bound to a 53-
Subsequent experiments showed that TRAP can also bind its target sequence in the leader region of mature mRNAs, blocking access of the ribosome to the Shine-
723
Autoinducer Analysis Reveals Possibilities for Treating Cholera
Higgins, D.A., M.E. Pomianek, C.M. Kraml, R.K. Taylor, M.F. Semmelhack, and B.L. Bassler. 2007. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450:883–
Many single-
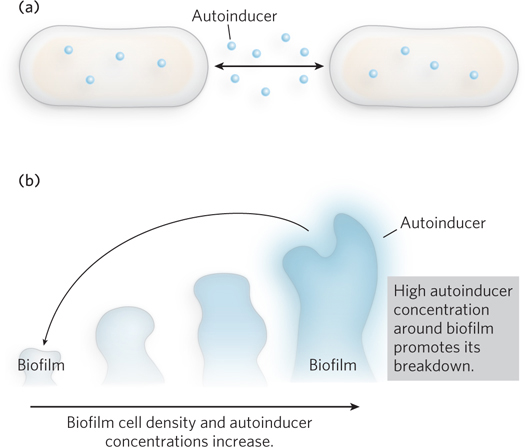
Bonnie Bassler’s laboratory at Princeton has investigated the molecular details of quorum sensing in V. cholerae. The bacterium uses quorum sensing to control its production of virulence factors and its ability to grow as a biofilm, a contiguous layer of cells. At low cell density, V. cholerae expresses virulence factors and forms a biofilm. As cell density increases, two autoinducers increase in concentration until they repress both virulence factor expression and biofilm formation (Figure 2b). Synthesized by autoinducer synthase enzymes, the small-
Information from both types of autoinducers is transmitted through the protein LuxO, which in turn controls the level of HapR, a transcription factor that controls the expression of many other genes. At low cell density, in the absence of autoinducers, HapR is not produced, virulence factors are expressed, and biofilms form. Because HapR is required for the expression of genes that cause the cells to luminesce, the cells are not bioluminescent. This provides an easy way for researchers to determine whether HapR is turned on. At high cell density, autoinducers increase and bind to LuxO, leading to the production of HapR. HapR represses the genes for virulence factor production and biofilm formation, while activating expression of the bioluminescence genes. The end result is that the production of autoinducers and activation of quorum sensing terminate virulence in V. cholerae cells growing in crowded conditions.
Bassler and colleagues cloned the autoinducer synthase genes and introduced them into E. coli. Because E. coli is not sensitive to the V. cholerae autoinducers, the signaling molecules were produced and secreted in large amounts without interfering with cell growth. The experimenters could then purify the autoinducers and determine their chemical structure by nuclear magnetic resonance spectroscopy (NMR). The NMR method enables exact determination of the chemical structure of the autoinducers, because it provides information about the chemical environment of each proton in the molecule. Bassler’s group has also developed a method for chemically synthesizing these autoinducers—
724