SOLVED PROBLEMS
SOLVED PROBLEM 1. Two Drosophila flies that had normal (transparent, long) wings were mated. In the progeny, two new phenotypes appeared: dusky wings (having a semi-
|
Females |
Males |
|
179 transparent, long |
92 transparent, long |
|
58 transparent, clipped |
89 dusky, long |
|
|
28 transparent, clipped |
|
|
31 dusky, clipped |
a. Provide a chromosomal explanation for these results, showing chromosomal genotypes of parents and of all progeny classes under your model.
b. Design a test for your model.
Solution
a. The first step is to state any interesting features of the data. The first striking feature is the appearance of two new phenotypes. We encountered the phenomenon in Chapter 2, where it was explained as recessive alleles masked by their dominant counterparts. So, first, we might suppose that one or both parental flies have recessive alleles of two different genes. This inference is strengthened by the observation that some progeny express only one of the new phenotypes. If the new phenotypes always appeared together, we might suppose that the same recessive allele determines both.
However, the other striking feature of the data, which we cannot explain by using the Mendelian principles from Chapter 2, is the obvious difference between the sexes: although there are approximately equal numbers of males and females, the males fall into four phenotypic classes, but the females constitute only two. This fact should immediately suggest some kind of sex-
Having done this partial analysis, we see that only the inheritance of wing transparency is associated with sex. The most obvious possibility is that the alleles for transparent (D) and dusky (d) are on the X chromosome, because we have seen in Chapter 2 that gene location on this chromosome gives inheritance patterns correlated with sex. If this suggestion is true, then the parental female must be the one sheltering the d allele, because, if the male had the d, he would have been dusky, whereas we were told that he had transparent wings. Therefore, the female parent would be D/d and the male D. Let’s see if this suggestion works: if it is true, all female progeny would inherit the D allele from their father, and so all would be transparent winged, as was observed. Half the sons would be D (transparent) and half d (dusky), which also was observed.
So, overall, we can represent the female parent as D/d; L/l and the male parent as D; L/l. Then the progeny would be
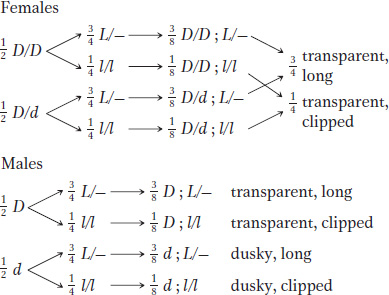
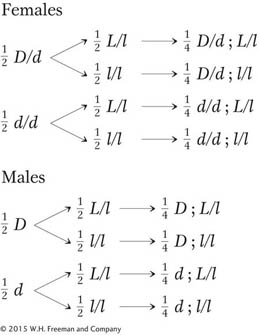
b. Generally, a good way to test such a model is to make a cross and predict the outcome. But which cross? We have to predict some kind of ratio in the progeny, and so it is important to make a cross from which a unique phenotypic ratio can be expected. Notice that using one of the female progeny as a parent would not serve our needs: we cannot say from observing the phenotype of any one of these females what her genotype is. A female with transparent wings could be D/D or D/d, and one with long wings could be L/L or L/l. It would be good to cross the parental female of the original cross with a dusky, clipped son, because the full genotypes of both are specified under the model that we have created. According to our model, this cross is
D/d; L/l × d; l/l
From this cross, we predict
SOLVED PROBLEM 2. Consider three yellow, round peas, labeled A, B, and C. Each was grown into a plant and crossed with a plant grown from a green wrinkled pea. Exactly 100 peas issuing from each cross were sorted into phenotypic classes as follows:

What were the genotypes of A, B, and C? (Use gene symbols of your own choosing; be sure to define each one.)
Solution
Notice that each of the crosses is
yellow, round × green, wrinkled → progeny
Because A, B, and C were all crossed with the same plant, all the differences between the three progeny populations must be attributable to differences in the underlying genotypes of A, B, and C.
You might remember a lot about these analyses from the chapter, which is fine, but let’s see how much we can deduce from the data. What about dominance? The key cross for deducing dominance is B. Here, the inheritance pattern is
yellow, round × green, wrinkled → all yellow, round
So yellow and round must be dominant phenotypes because dominance is literally defined by the phenotype of a hybrid. Now we know that the green, wrinkled parent used in each cross must be fully recessive; we have a very convenient situation because it means that each cross is a test-
Turning to the progeny of A, we see a 1:1 ratio for yellow to green. This ratio is a demonstration of Mendel’s first law (equal segregation) and shows that, for the character of color, the cross must have been heterozygote × homozygous recessive. Letting Y represent yellow and y represent green, we have
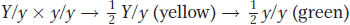
For the character of shape, because all the progeny are round, the cross must have been homozygous dominant χ homozygous recessive. Letting R represent round and r represent wrinkled, we have
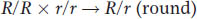
Combining the two characters, we have
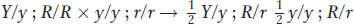
Now cross B becomes crystal clear and must have been
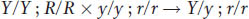
because any heterozygosity in pea B would have given rise to several progeny phenotypes, not just one.
What about C? Here, we see a ratio of 50 yellow:50 green (1:1) and a ratio of 49 round:51 wrinkled (also 1:1). So both genes in pea C must have been heterozygous, and cross C was
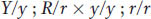
which is a good demonstration of Mendel’s second law (independent assortment of different genes).
How would a geneticist have analyzed these crosses? Basically, the same way that we just did but with fewer intervening steps. Possibly something like this: “yellow and round dominant; single-