Immunoglobulin diversity results from DNA rearrangements and other mutations
Each mature B cell makes antibodies targeted to only one single antigen. And there are millions of possible antigens to which a human can be exposed. How can the genome encode enough different antibodies to protect the body against all the possible pathogens? Although there are millions of possible amino acid sequences in immunoglobulins, there are not millions of different immunoglobulin genes. It turns out that instead of a single gene encoding each complete immunoglobulin, the genome of the differentiating B cell has multiple different coding regions for each domain of the protein, and diversity is generated by putting together different combinations of these regions. Shuffling of this genetic deck generates the enormous immunological diversity that characterizes each individual mammal.
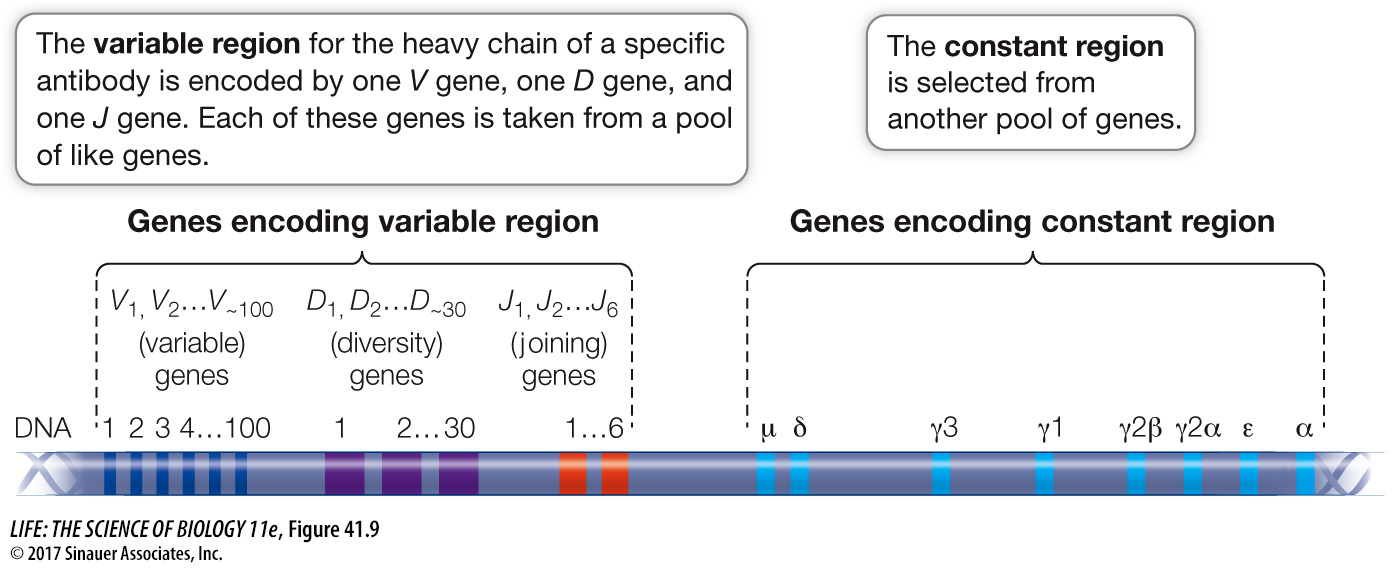
Each gene encoding an immunoglobulin chain is in reality a “supergene” assembled by means of genetic recombination from several clusters of smaller genes scattered along part of a chromosome (Figure 41.9). Every cell in the body has hundreds of immunoglobulin genes located in separate clusters that are potentially capable of participating in the synthesis of both the variable and constant regions of immunoglobulin chains. In most body cells and tissues, these genes remain intact and separated from one another. But during B cell development, these genes are cut out, rearranged, and joined together in DNA recombination events. One gene from each cluster is chosen randomly for joining, and the others are deleted. In the case of one multigene set, the J genes, some of the extra sequences are removed by RNA splicing (Figure 41.10).
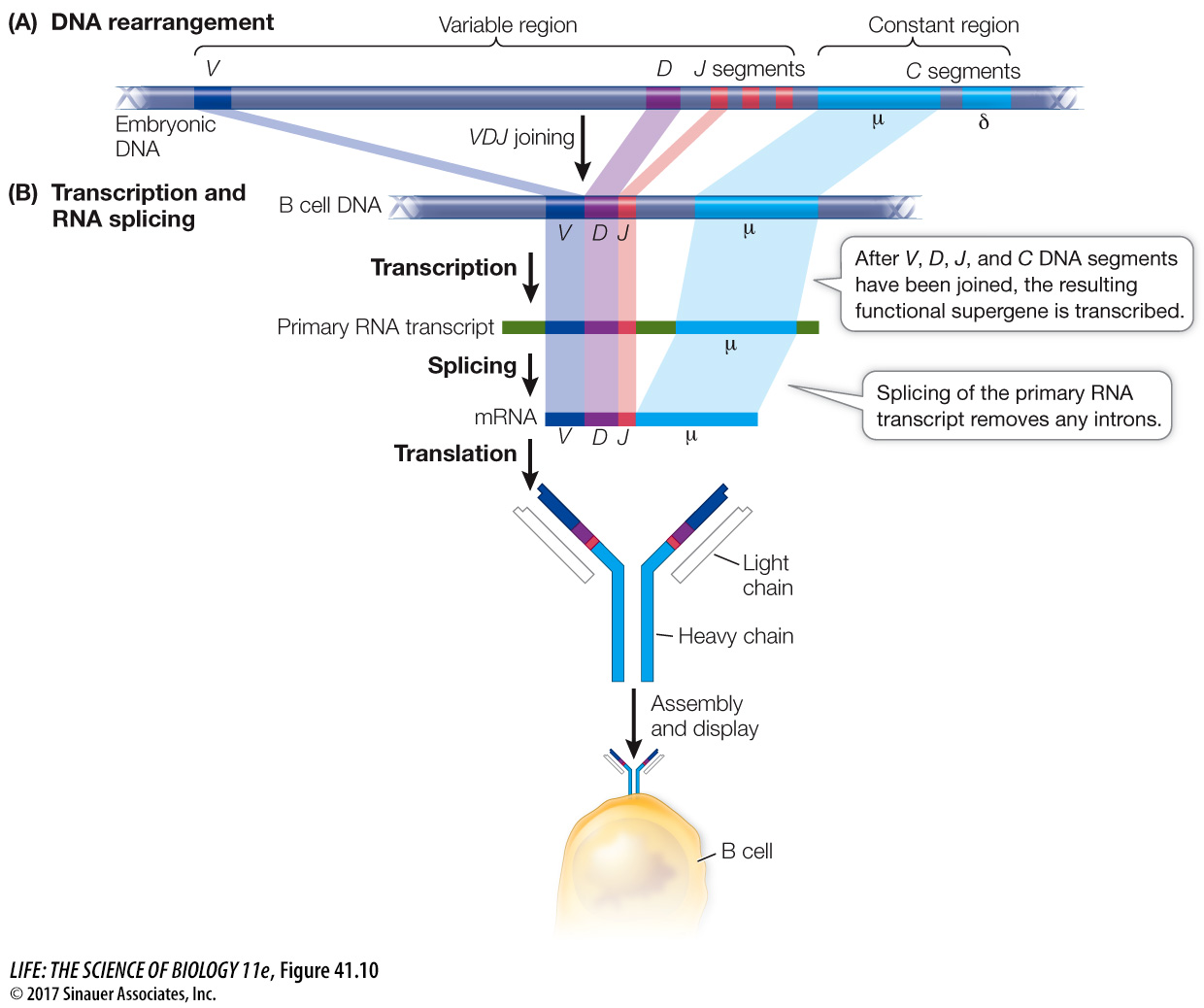
In this manner, a unique immunoglobulin supergene is assembled from randomly selected “parts.” Each B cell precursor assembles two supergenes, one for a specific heavy chain and the other, assembled independently, for a specific light chain. This remarkable example of irreversible cell differentiation generates an enormous diversity of immunoglobulins from the same genome. It is a major exception to the generalization that all somatic cells derived from the fertilized egg have identical DNA.
Figure 41.9 illustrates the gene families that encode the constant and variable regions of the heavy chain in mice. Multiple genes encode each of the three parts of the variable region: 100 V, 30 D, and 6 J genes. Each B cell randomly selects one gene from each of these clusters to make the final coding sequence (VDJ) of the heavy-
100 V × 30 D × 6 J = 18,000 possible combinations
Now consider that the light chains are similarly constructed, with a similar amount of diversity made possible by random recombination. If we assume that the degree of potential light-
18,000 different light chains × 18,000 different heavy chains = 324 million possibilities!
Other mechanisms generate even more diversity:
When the DNA sequences that encode the V, D, and J regions are rearranged so that they are next to one another, the recombination event is not precise, and errors occur at the junctions. This imprecise recombination can create *frame-
shift mutations , generating new codons at the junctions, with resulting amino acid changes.After the DNA sequences are cut and before they are rejoined, the enzyme terminal transferase often adds some nucleotides to the free ends of the DNA pieces. These additional bases create insertion mutations.
There is a relatively high spontaneous mutation rate in immunoglobulin genes. Once again, this process creates many new alleles and adds to antibody diversity.
*connect the concepts When they occur in the coding regions of proteins, point mutations can cause silent, missense, nonsense, or frame-
When we include these possibilities with the millions of combinations that can be made by random DNA rearrangements, it is not surprising that the immune system can mount a response to almost any natural or artificial substance.
Once the DNA rearrangements are completed, each supergene is transcribed and then translated to produce an immunoglobulin light chain or heavy chain. These chains combine to form an active immunoglobulin protein.
Animation 41.3 A B Cell Builds an Antibody