13.4 Targeting of Proteins to Mitochondria and Chloroplasts
In the remainder of this chapter, we examine how proteins synthesized on cytosolic ribosomes are sorted to the discrete organelles: mitochondria, chloroplasts, peroxisomes, and the nucleus (see Figure 13-1). Mitochondria and chloroplasts are closely related organelles that contain an internal lumen, called the matrix, that is surrounded by a double membrane. In contrast, peroxisomes, which we discuss next, are bounded by a single membrane and have a single luminal matrix compartment. The mechanism of protein transport into and out of the nucleus, which differs in many respects from sorting to other organelles, is discussed in the last section of the chapter.
In addition to being bounded by two membranes, mitochondria and chloroplasts share similar types of electron-transporting proteins and use an F-class ATPase to synthesize ATP (see Figure 12-30). Remarkably, these characteristics are shared by gram-negative bacteria. Also like bacterial cells, mitochondria and chloroplasts contain their own DNA, which encodes organelle rRNAs, tRNAs, and some proteins (see Chapter 8). Moreover, growth and division of mitochondria and chloroplasts are not coupled to nuclear division. Rather, these organelles grow by the incorporation of cellular proteins and lipids, and new organelles form by division of preexisting organelles. These numerous similarities to free-living bacterial cells have led to the understanding that mitochondria and chloroplasts arose when bacteria were incorporated into ancestral eukaryotic cells, forming endosymbiotic organelles (see Figure 12-7). The sequence similarity of the many membrane translocation proteins shared by mitochondria, chloroplasts, and bacteria provides the most striking evidence for this ancient evolutionary relationship. In this section, we examine these membrane translocation proteins in detail.
Page 609
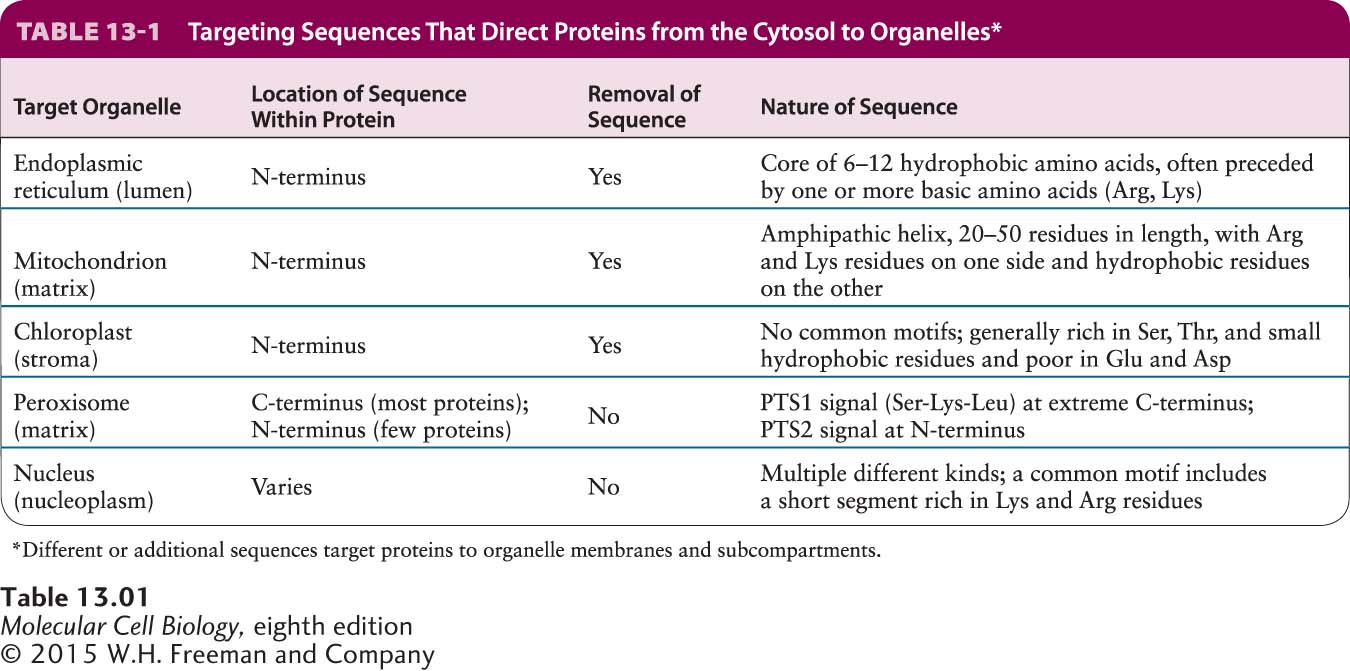
Proteins encoded by mitochondrial DNA or chloroplast DNA are synthesized on ribosomes within the organelles and directed to the correct subcompartment within the parent organelle immediately after synthesis. However, the majority of proteins located in mitochondria and chloroplasts are encoded by genes in the nucleus and are imported into the organelles after their synthesis in the cytosol. Apparently, as eukaryotic cells evolved over a billion years, much of the genetic information from the ancestral bacterial DNA in these endosymbiotic organelles moved, by an unknown mechanism, to the nucleus. Precursor proteins synthesized in the cytosol that are destined for the matrix of mitochondria or the equivalent space in chloroplasts, the stroma, usually contain specific N-terminal targeting sequences that specify binding to receptor proteins on the organelle surface. Generally these sequences are cleaved once the protein reaches the matrix or stroma. Clearly these targeting sequences are similar in their location and general function to the signal sequences that direct nascent proteins to the ER lumen. Although the three types of signals share some sequence features, their specific sequences differ considerably, as summarized in Table 13-1.
In both mitochondria and chloroplasts, protein import requires energy and occurs at points where the outer and inner organelle membranes are in direct contact. Because mitochondria and chloroplasts contain multiple membranes and membrane-limited spaces, the sorting of many proteins to their correct locations requires the sequential action of two targeting sequences and two membrane-bound translocation systems: one to direct the protein into the organelle and the other to direct it into the correct organelle subcompartment or membrane. As we will see, the mechanisms for sorting various proteins to mitochondria and chloroplasts are related to some of the mechanisms discussed previously.