Normal Yeast Genes Can Be Replaced with Mutant Alleles by Homologous Recombination
Modifying the genome of the yeast S. cerevisiae is particularly easy for two reasons: yeast cells readily take up exogenous DNA under certain conditions, and the introduced DNA is efficiently exchanged for the homologous chromosomal site in the recipient cell. This specific, targeted recombination of identical stretches of DNA allows any gene in yeast chromosomes to be replaced with a mutant allele. (As we saw in Section 6.1, recombination between homologous chromosomes also occurs naturally during meiosis.)
In one popular method for disrupting yeast genes in this fashion, PCR is used to generate a disruption construct containing a selectable marker, which is subsequently transfected into yeast cells. As shown in Figure 6-36a, the two primers for PCR amplification of the selectable marker are each designed to include about 20 nucleotides identical to sequences flanking the yeast gene to be replaced. The resulting amplified construct comprises the selectable marker (e.g., the kanMX gene, which, like neor, confers resistance to G-
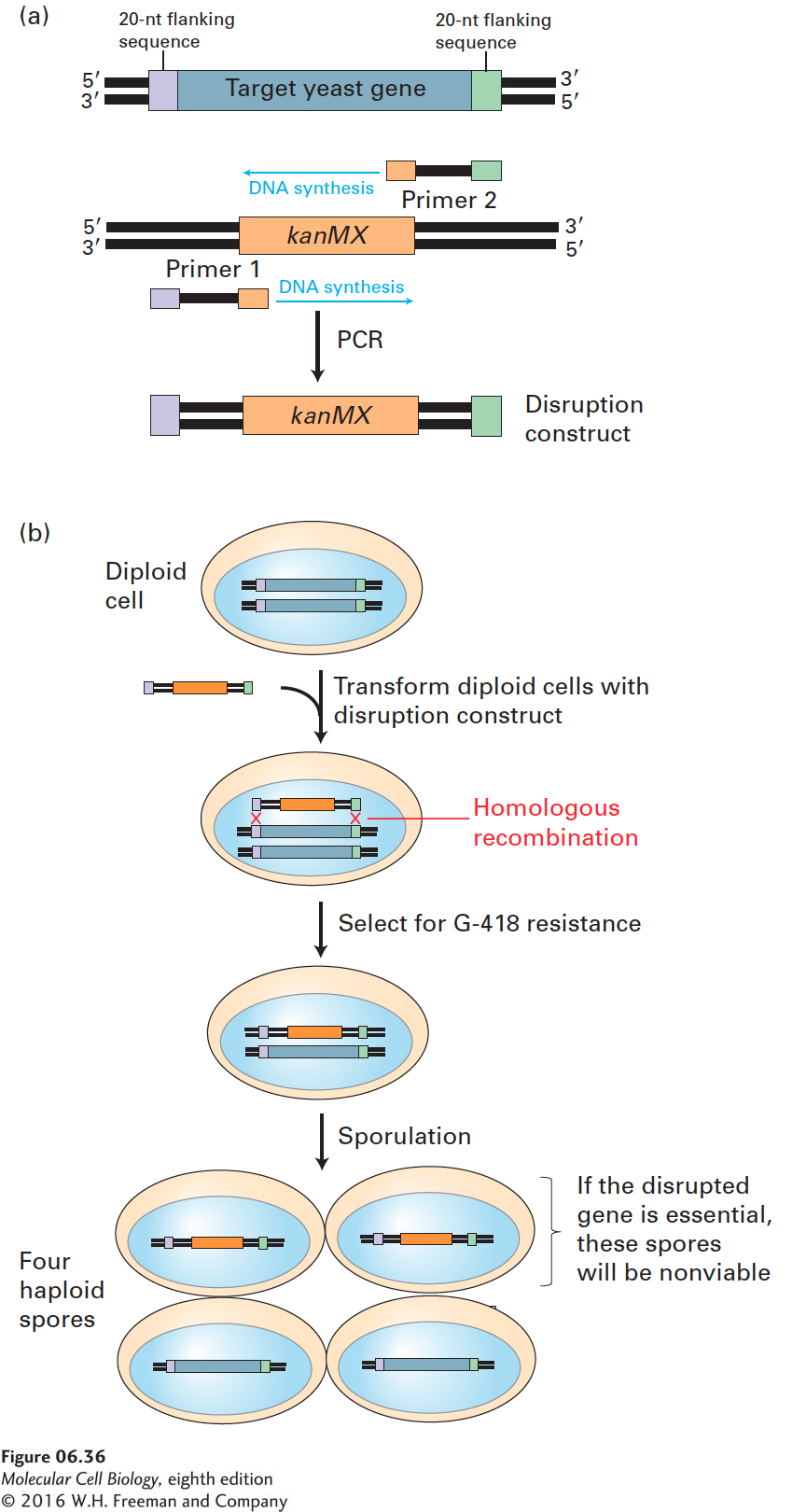
Disruption of genes by this method is proving particularly useful in assessing the roles of proteins identified by analysis of the entire genomic sequence of S. cerevisiae (see Chapter 8). Each of the approximately 6000 genes has been disrupted with the kanMX construct in diploids, and gene disruptions in haploid spores have also been produced. These analyses have shown that about 4500 of the 6000 yeast gene disruptions can reside in viable haploid spores, revealing an unexpectedly large number of apparently nonessential genes. In some cases, disruption of a particular gene may give rise to subtle defects that do not compromise the viability of yeast cells growing under laboratory conditions. Alternatively, cells carrying a disrupted gene may be viable because of the operation of backup or compensatory pathways. To investigate this possibility, yeast geneticists are currently testing all possible double-