Molecular data complement comparative morphology in reconstructing phylogenetic history.
Trees can be built using anatomical features, but increasingly tree construction relies on molecular data. The amino acids at particular positions in the primary structure of a protein can be used, as can the nucleotides at specific positions along a strand of DNA.
From genealogy to phylogeny, tracing mutations in DNA or RNA sequences has revolutionized the reconstruction of historical genetic connections. Whether we are tracing the paternity of the children of Sally Hemings, mistress of Thomas Jefferson, identifying the origin of a recent cholera epidemic in Haiti, or placing baleen whales near the hippopotamus family in a phylogenetic tree of mammals, molecular data are a rich source of phylogenetic insight.
There is nothing about molecular data that provides a better record of history than does anatomical data; molecular data simply provide more details because there are more characters that can vary among the species. A sequence of DNA with hundreds or thousands of nucleotides can represent that many characters, as opposed to the tens of characters usually visible in morphological studies. Indeed, for microbes and viruses there is very little morphology available, so molecular information is critical for phylogenetic reconstruction. Once a gene or other stretch of DNA or RNA is identified that seems likely (based on previous studies of other species) to vary among the species to be studied, sequences are obtained and aligned to identify homologous nucleotide sites. Analyses of this kind commonly involve comparisons of sequences of about 1000 nucleotides from one or more genes. Increasingly, though, the availability of whole-
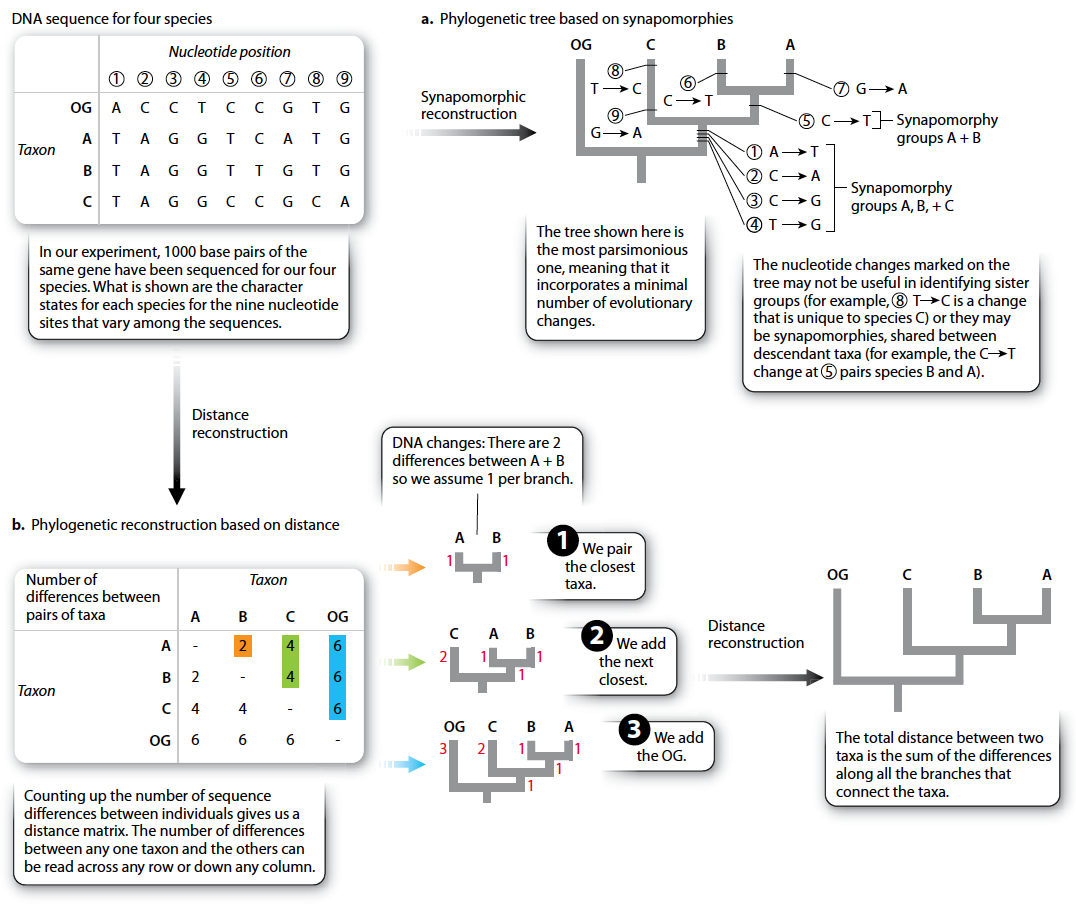
The process of using molecular data is conceptually similar to the process described earlier for morphological data. Through comparison to an outgroup, we can identify derived and ancestral molecular characters (whether DNA nucleotides or amino acids in proteins) and generate the phylogeny on the basis of synapomorphies as before.
An alternative method of reconstruction is based on overall similarity rather than synapomorphies. Here, the premise is simple: The descendants of a recent common ancestor will have had relatively little time to evolve differences, whereas the descendants of an ancient common ancestor have had a lot of time to evolve differences. Thus, the extent of similarity (or distance) indicates how recently two groups shared a common ancestor.
Underpinning this approach is the assumption that the rate of evolution is constant. (Otherwise, a pair of taxa with a recent common ancestor could be more different than expected because of an unusually fast rate of evolution.) This rate-
Molecular data are often combined with morphological data, and each can also serve as an independent assessment of the other. Not surprisingly, results from analyses of each kind of data are commonly compatible, at least for plants and animals rich in morphological characters.
The single largest library of taxonomic information is GenBank, the National Institutes of Health’s genetic data storage facility. As of this writing, GenBank gives users access to more than 100 billion observations (mostly nucleotides) collected under more than 430,000 taxonomic names. A growing internet resource is the Encyclopedia of Life, which is gathering additional biological information about species, including ecology, geographic distributions, photographs, and sounds in pages for individual species that are easy to navigate. Another web resource, the Tree of Life, provides information on phylogenetic trees for many groups of organisms.