Chapter 4 Summary
Core Concepts Summary
4.1 Proteins are linear polymers of amino acids that form three-
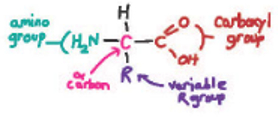
An amino acid consists of an α carbon connected by covalent bonds to an amino group, a carboxyl group, a hydrogen atom, and a side chain or R group. page 70
There are 20 common amino acids that differ in their R groups. Amino acids are categorized by the chemical properties of their R groups—
Amino acids are connected by peptide bonds to form proteins. page 72
The primary structure of a protein is its amino acid sequence. The primary structure determines how a protein folds, which in turn determines how it functions. page 73
The secondary structure of a protein results from the interactions of nearby amino acids. Examples include the α helix and β sheet. page 73
The tertiary structure of a protein is its three-
Some proteins are made up of several polypeptide subunits; this group of subunits is the protein’s quaternary structure. page 76
Chaperones help some proteins fold properly. page 76
4.2 Translation is the process by which the sequence of bases in messenger RNA specifies the order of successive amino acids in a newly synthesized protein.
Translation requires many cellular components, including ribosomes, tRNAs, and proteins. page 77
Ribosomes are composed of a small and a large subunit, each consisting of RNA and protein; the large subunit contains three tRNA-
An mRNA transcript of a gene has three possible reading frames composed of three-
tRNAs have an anticodon that base pairs with the codon in the mRNA and carries a specific amino acid. page 78
Aminoacyl tRNA synthetases attach specific amino acids to tRNAs. page 79
The genetic code defines the relationship between the three-
The genetic code is redundant in that many amino acids are specified by more than one codon. page 81
Translation consists of three steps: initiation, elongation, and termination. page 81
4.3 Proteins evolve through mutation and selection and by combining functional units.
A protein family is a group of proteins that are structurally and functionally related. page 86
There are far fewer protein families than the total number of possible proteins because the probability that a random sequence of amino acids will fold properly to carry out a specific function is very small. page 86
A region of a protein that folds in a particular way and that carries out a specific function is called a folding domain. page 86
Proteins evolve by combining different folding domains. page 86
Proteins also evolve by changes in amino acid sequence, which occurs by mutation and selection. page 87
Self-Assessment
Draw one of the 20 amino acids and label the amino group, the carboxyl group, the R group (side chain), and the α carbon.
Self-Assessment 1 Answer
Briefly, the carboxyl group and α carbon are the same for all amino acids (see Fig. 4.2). The amino group is the same for almost all the amino acids, with proline being the exception. The variety of function and form between the amino acids is mainly due to the different side chains (R groups).
Name four major groups of amino acids, categorized by the properties of their R groups. Explain how the chemical properties of each group affect protein shape.
Self-Assessment 2 Answer
Amino acids can be categorized into five main groups based on the properties of their side chains: The first are the hydrophobic amino acids, “water-
fearing,” whose side chains are nonpolar, usually found buried in the interior of the folded proteins, and typically form bonds with other hydrophobic amino acids or solvents (e.g., valine). In the second group are the hydrophilic amino acids, “water- loving,” that have polar side chains, usually found on the outside surface of folded proteins, and typically form bonds with other hydrophilic amino acids or water. Their charge allows them to interact with other proteins and macromolecules. Hydrophilic amino acids are also broken up into two groups: basic amino acids with side chains that are positively charged at intracellular pH (e.g., lysine) and acidic amino acids with side chains that are negatively charged (e.g., aspartic acid). The fifth group are “special” amino acids, whose unique structures and chemical properties have significant effects on higher levels of protein structure because they allow more or less flexibility around the peptide bond (glycine and proline), or covalent interactions between side chains (cysteine). Describe how peptide bonds, hydrogen bonds, ionic bonds, disulfide bridges, and noncovalent interactions (van der Waals forces and the hydrophobic effect) define a protein’s four levels of structure.
Self-Assessment 3 Answer
The way in which amino acids interact and bond in a polypeptide chain is important for the structure and function of the protein.
Peptide bonds are important in maintaining the primary structure of a polypeptide chain. These bonds form between the carboxyl group of one amino acid and the amino group of the next amino acid in the chain. Note that these bonds are typically found between amino acid residues in the polypeptide.
Hydrogen bonds are important in maintaining the secondary structure of the polypeptide chain. These bonds form between the oxygen in the carbonyl group (C=O) of one peptide bond and the hydrogen in the amide group (NH) of another. This allows regions of the polypeptide to interact with itself and fold. Two common types of secondary structure formed by hydrogen bonding are α helices and β sheets (Figs. 4.6 and 4.7). Note that in terms of secondary structure, these bonds are found between groups in the polypeptide backbone.
There are four groups of bonds or interactions important in creating tertiary and quaternary structure. The first group is the ionic bonds that form between a negative charge and a positive charge. For example, an ionic bond would form between a basic amino acid and an acidic amino acid because they have oppositely charged side chains. These bonds can occur between amino acids that are far apart in the polypeptide chain, thus creating loops and bends in the overall structure. The second group is the hydrogen bonds that form between the oxygen of one amino acid’s side chain and the hydrogen of another amino acid’s side chain. The third group is the disulfide bridges. These covalent bonds form between two cysteine residues in the same polypeptide chain, or between two cysteines in two different chains. Note that when discussing tertiary structure, these bonds are found between different side chains. The fourth group important to maintaining tertiary and quaternary structure is noncovalent interactions that include van der Waals forces and hydrophobic interactions that maintain interactions with different domains of the protein and result in a protein’s specific shape.
Explain how the order of amino acids determines the way in which a protein folds.
Self-Assessment 4 Answer
The order of amino acids in the polypeptide chain determines the way in which proteins fold because of the various interactions and bonds formed between the amino acids. These interactions, depending on the type and location, will give rise to a specific secondary and tertiary structure. More often than not, these structures must be perfectly arranged for the protein to function. Thus, it is important that the polypeptide chain be correctly ordered to result in the specific structure of that particular protein.
Explain the relationship between protein folding and protein function.
Self-Assessment 5 Answer
In many cases, the ability of a protein to perform its function is dependent upon the protein being in the correct conformation. For example, many enzymes bind to their substrate through specific interactions between their active sites and a site on the substrate. If a mutation causes an amino acid change in the gene resulting in the enzyme having a different shape or different chemical property (e.g., negatively charged instead of polar), it may no longer be able to bind to its substrate and perform its activity.
Describe the relationship between the template strand of DNA, the codons in mRNA, anticodons in tRNA, and amino acids.
Self-Assessment 6 Answer
The codons of mRNA are groups of three nucleotides that code for a particular amino acid. Each is transcribed from the template strand of DNA according to the normal rules of base pairing (but in RNA, U replaces T). The sequence of the codons in the mRNA gives rise to the order of the resulting amino acid polypeptide chain. The codons are translated by tRNAs. The sequence of each tRNA includes a group of three nucleotides called an anticodon that is complementary in sequence and thus can recognize and bind to a specific codon in the mRNA. Because of the complementary and antiparallel nature of nucleic acid structures, an anticodon in a tRNA has the same 3' to 5' sequence as the template DNA, except with U’s instead of T’s. Each tRNA is also bound to a specific amino acid, affiliated with a particular anticodon/codon pair, on the 3' end of the molecule. When the mRNA is being “read” through the ribosome, the order of the amino acids in the polypeptide chain is dependent upon the sequential interaction of the mRNA codon with the correct tRNA anticodon/amino acid pair. See Fig. 4.15.
Describe the steps of translation initiation, elongation, and termination.
Self-Assessment 7 Answer
Translation of mRNA by ribosomes can be divided into three processes:
Initiation:Initiation factors bind to the 5' cap of the mRNA (in eukaryotic cells) or at the Shine‒Dalgarno sequence (for prokaryotes), and recruit the small subunit of the ribosome and a tRNA charged with methionine. This complex then moves along the mRNA until it finds a start codon (AUG, coding for methionine). The large ribosomal subunit then joins the complex and causes the initiation factors to be released. The tRNAMet is then bound in the P site of the ribosome. The next tRNA, determined by the codon of the mRNA, binds in the A site of the ribosome. This elicits a coupled reaction in which the bond between the Met and its tRNA is broken and a new bond is formed between the carboxyl group of the Met and the amino group of the next amino acid (a peptide bond). The ribosome complex then slides to the next codon on the mRNA, shifting the now uncharged tRNAMet to the E site, where it is released from the ribosome complex, and the peptide-
bearing tRNA to the P site. The A site is now free for the next charged tRNA. Elongation:The ribosome continues in this fashion, shifting down the mRNA one codon at a time, adding amino acids to the growing peptide chain. Elongation factors provide the energy needed for these reactions to happen.
Termination:When the ribosome complex comes across a stop codon (UAA, UAG, or UGA), a protein release factor binds in the A site of the ribosome and causes the bond between the polypeptide chain and the last tRNA to break. Once the polypeptide chain is released, the ribosomal subunits disassociate from the mRNA and each other and translation is complete.
Name and describe two ways that proteins can acquire new functions in the course of evolution.
Self-Assessment 8 Answer
Two ways in which proteins can acquire new functions through the course of evolution are (1) mutation and selection and (2) combining different folding domains.
(1) Mutation and selection: The sequence of the amino acids in the polypeptide chain is important for the proper folding, and ultimately the function, of the protein. If the sequence is altered by a mutation that changes a codon to specify for a different amino acid, this could affect the function of the protein and whether it is selected for in the population. A mutation that leads to a nonfunctioning protein will most likely lead to the impaired survival and reproductive ability of that organism and will thus be eventually eliminated from the population. A neutral mutation that does not impair or improve protein function will likely remain in the population because those organisms will survive and reproduce at normal levels. A mutation that improves the function of the protein, although rare, would give a selective advantage to that organism if it could survive and reproduce more successfully.
(2) Combining different folding domains: Form leads to function. If a gene gains a new folding domain by joining with a folding domain from another gene, for example, its product now has the additional function provided by that folding domain. If this function is beneficial or benign to the protein and ultimately to the survival and reproductive ability of the organism, the new gene, and therefore protein, will be maintained in the population.